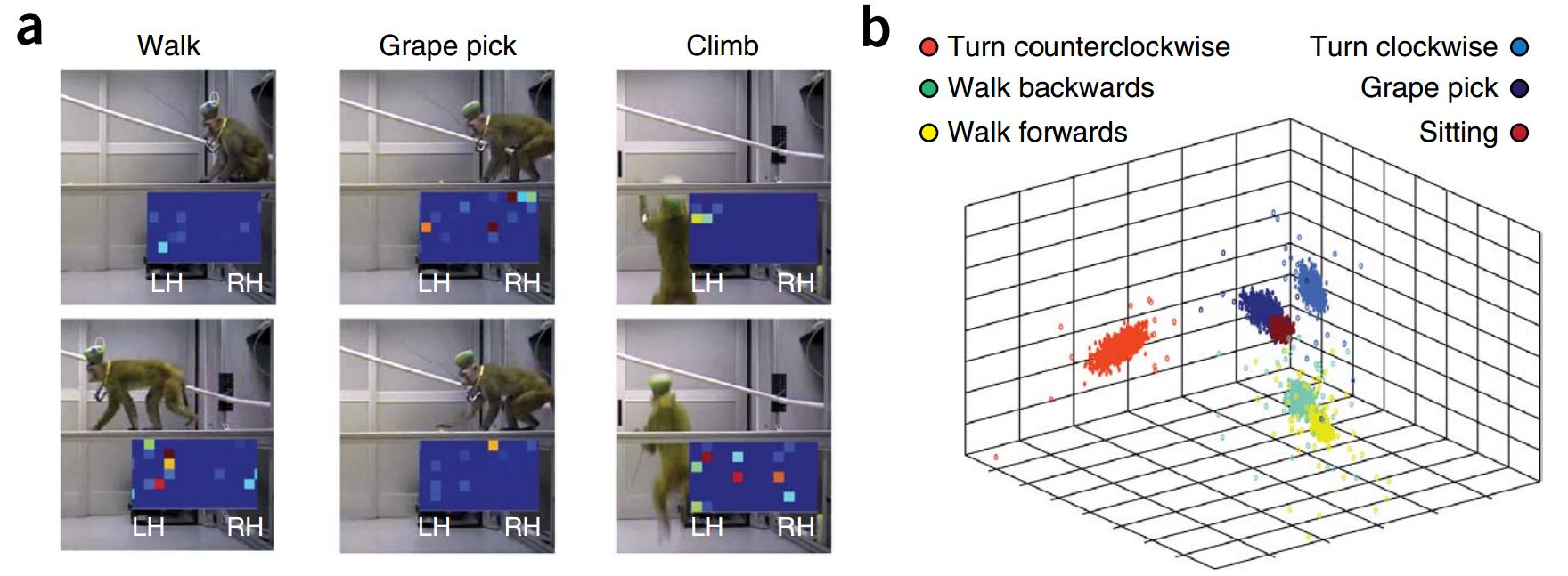
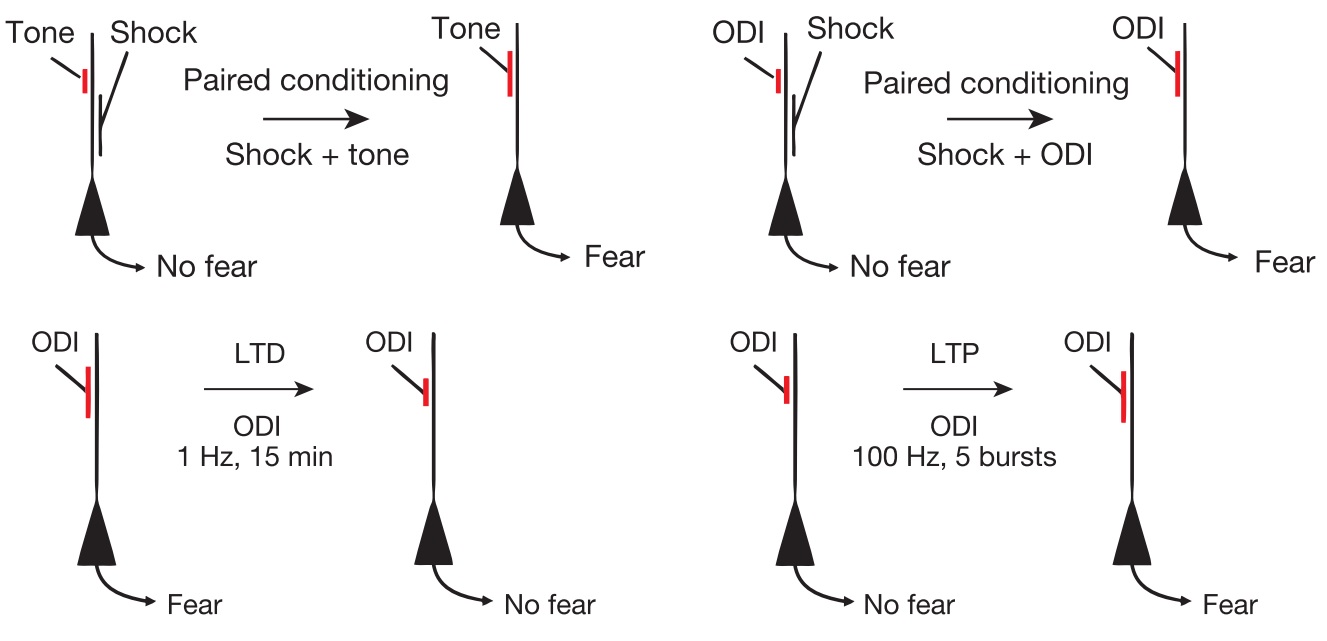
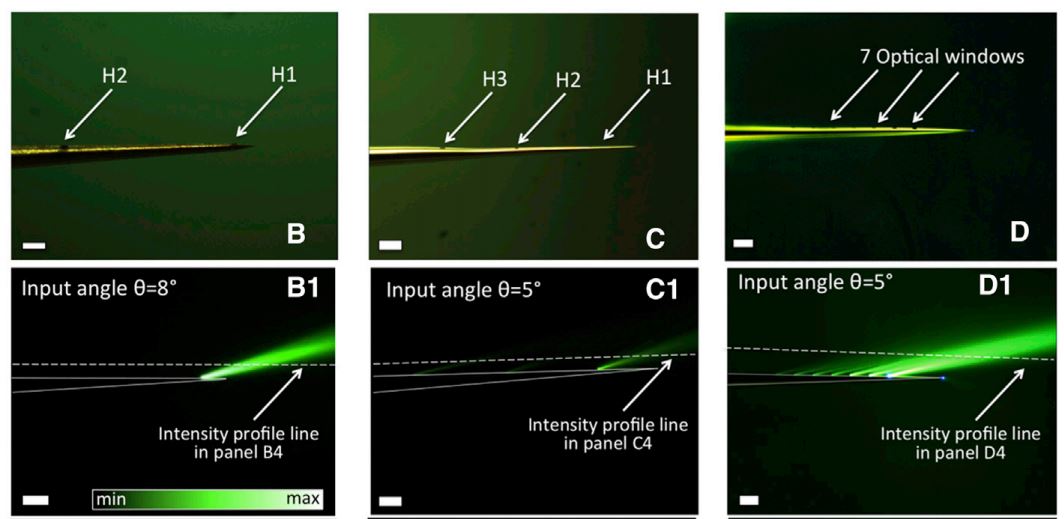
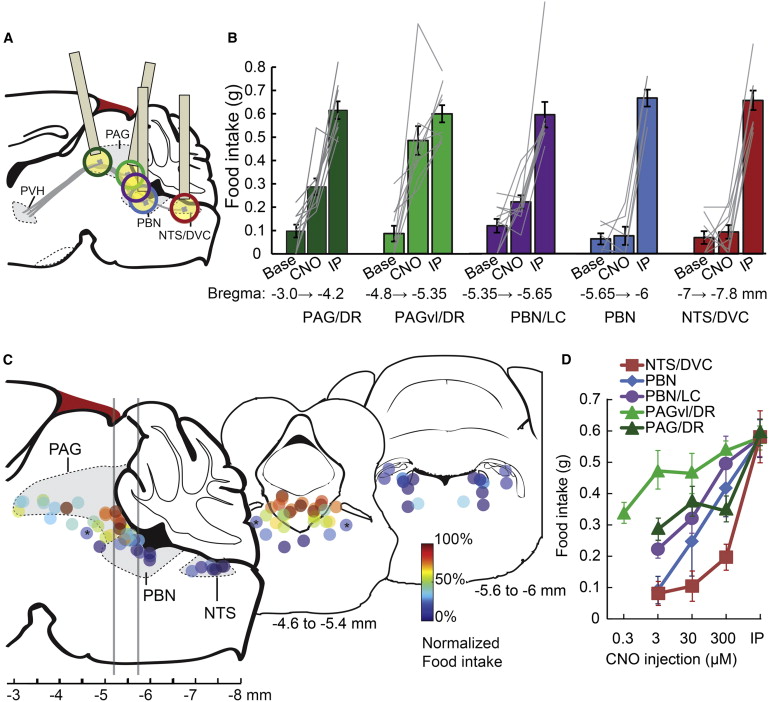
B Srinivasa Reddy and Biswanath N Chatterji. An fft-based technique
for translation, rotation, and scale-invariant image registration. IEEE
transactions on image processing, 5(8):1266–1271, 1996.
Philippe Thevenaz, Urs E Ruttimann, and Michael Unser. A pyramid
approach to subpixel registration based on intensity. Image Processing,
IEEE Transactions on, 7(1):27–41, 1998.
Hassan Foroosh, Josiane B Zerubia, and Marc Berthod. Extension of
phase correlation to subpixel registration. IEEE transactions on image
processing, 11(3):188–200, 2002.
J N Kerr, D Greenberg, and F Helmchen. Imaging input and output
of neocortical networks in vivo. Proc Natl Acad Sci U S A, 102(39):
14063–14068, 2005. ISSN 0027-8424 (Print) 0027-8424. doi: 10.1073/pnas.
0506029102.
K V Kuchibhotla, S Wegmann, K J
Kopeikina, J Hawkes, N Rudinskiy, M L Andermann, T L Spires-Jones,
B J Bacskai, and B T Hyman. Neurofibrillary tangle-bearing neurons are
functionally integrated in cortical circuits in vivo. Proc Natl Acad Sci U S
A, 111(1):510–514, 2014. ISSN 0027-8424. doi: 10.1073/pnas.1318807111.
Simon P. Peron, Jeremy Freeman, Vijay Iyer, Caiying Guo, and Karel
Svoboda. A Cellular Resolution
Map of Barrel Cortex Activity during Tactile Behavior. Neuron, 86(3):
783–799, 2015. ISSN 10974199. doi: 10.1016/j.neuron.2015.03.027. URL
http://dx.doi.org/10.1016/j.neuron.2015.03.027.
Anne E Carpenter, Thouis R Jones, Michael R Lamprecht, Colin
Clarke, In Han Kang, Ola Friman, David A Guertin, Joo Han Chang,
Robert A Lindquist, Jason Moffat, et al. Cellprofiler: image analysis
software for identifying and quantifying cell phenotypes. Genome biology,
7(10):1–11, 2006.
Claire McQuin, Allen Goodman, Vasiliy Chernyshev, Lee Kamentsky,
Beth A Cimini, Kyle W Karhohs, Minh Doan, Liya Ding, Susanne M
Rafelski, Derek Thirstrup, et al. Cellprofiler 3.0: Next-generation image
processing for biology. PLoS biology, 16(7):e2005970, 2018.
Michael R Lamprecht, David M Sabatini, and Anne E Carpenter.
Cellprofiler™: free, versatile software for automated biological image
analysis. Biotechniques, 42(1):71–75, 2007.
Eran A Mukamel, Axel Nimmerjahn, and Mark J Schnitzer. Automated
analysis of cellular signals from large-scale calcium imaging data. Neuron,
63(6):747–760, 2009.
Brian B Avants, Nick Tustison, Gang Song, et al. Advanced
normalization tools (ants). Insight j, 2(365):1–35, 2009.
Stefan Klein, Marius Staring, Keelin Murphy, Max A Viergever, and
Josien PW Pluim. Elastix: a toolbox for intensity-based medical image
registration. IEEE transactions on medical imaging, 29(1):196–205, 2009.
David S Greenberg and Jason ND Kerr. Automated correction of fast
motion artifacts for two-photon imaging of awake animals. Journal of
neuroscience methods, 176(1):1–15, 2009.
A Miri, K Daie,
R D Burdine, E Aksay, and D W Tank. Regression-based identification
of behavior-encoding neurons during large-scale optical imaging of neural
activity at cellular resolution. J Neurophysiol, 105(2):964–980, 2011. ISSN
1522-1598 (Electronic) 0022-3077 (Linking). doi: 10.1152/jn.00702.2010.
URL http://www.ncbi.nlm.nih.gov/pubmed/21084686.
Angela Bauch, Izabela Adamczyk, Piotr Buczek, Franz-Josef Elmer,
Kaloyan Enimanev, Pawel Glyzewski, Manuel Kohler, Tomasz Pylak,
Andreas Quandt, Chandrasekhar Ramakrishnan, et al. openbis: a flexible
framework for managing and analyzing complex data in biology research.
BMC bioinformatics, 12(1):1–19, 2011.
M Francis, X Qian, C Charbel, J Ledoux, J C Parker, and M S Taylor.
Automated region of interest analysis of dynamic Ca(2)+ signals in image
sequences. Am J Physiol Cell Physiol, 303(3):C236–43, 2012. ISSN
0363-6143. doi: 10.1152/ajpcell.00016.2012.
Chris Allan, Jean-Marie Burel, Josh Moore, Colin Blackburn, Melissa
Linkert, Scott Loynton, Donald MacDonald, William J Moore, Carlos
Neves, Andrew Patterson, et al. Omero: flexible, model-driven data
management for experimental biology. Nature methods, 9(3):245–253, 2012.
Ferran Diego, Susanne Reichinnek, Martin Both, and Fred A Hamprecht.
Automated identification of neuronal activity from calcium imaging by
sparse dictionary learning. In Biomedical Imaging (ISBI), 2013 IEEE 10th
International Symposium on, pages 1058–1061. IEEE, 2013.
Jakub Tomek, Ondrej Novak, and Josef Syka. Two-photon processor and
seneca: a freely available software package to process data from two-photon
calcium imaging at speeds down to several milliseconds per frame. Journal
of neurophysiology, 110(1):243–256, 2013.
Eftychios A Pnevmatikakis, Yuanjun Gao, Daniel Soudry, David Pfau,
Clay Lacefield, Kira Poskanzer, Randy Bruno, Rafael Yuste, and Liam
Paninski. A structured matrix factorization framework for large scale
calcium imaging data analysis. arXiv preprint arXiv:1409.2903, 2014.
Ryuichi Maruyama, Kazuma Maeda, Hajime Moroda, Ichiro Kato,
Masashi Inoue, Hiroyoshi Miyakawa, and Toru Aonishi. Detecting cells
using non-negative matrix factorization on calcium imaging data. Neural
Netw, 55:11–19, mar 2014. ISSN 0893-6080. doi: 10.1016/j.neunet.2014.
03.007. URL http://www.ncbi.nlm.nih.gov/pubmed/24705544.
Patrick Kaifosh, Jeffrey D Zaremba, Nathan B Danielson, and Attila
Losonczy. SIMA: Python software for analysis of dynamic fluorescence
imaging data. Frontiers in neuroinformatics, 8:80, 2014.
Dimitri Yatsenko, Jacob Reimer, Alexander S Ecker, Edgar Y Walker,
Fabian Sinz, Philipp Berens, Andreas Hoenselaar, R James Cotton,
Athanassios S Siapas, and Andreas S Tolias. Datajoint: managing big
scientific data using matlab or python. BioRxiv, page 031658, 2015.
Jeffery L Teeters, Keith Godfrey, Rob Young, Chinh Dang, Claudia
Friedsam, Barry Wark, Hiroki Asari, Simon Peron, Nuo Li, and Adrien
Peyrache. Neurodata without borders: creating a common data format for
neurophysiology. Neuron, 88(4):629–634, 2015.
Marius Pachitariu, Carsen Stringer, Sylvia Schröder, Mario Dipoppa,
L Federico Rossi, Matteo Carandini, and Kenneth D Harris. Suite2p:
beyond 10,000 neurons with standard two-photon microscopy. Biorxiv,
page 061507, 2016.
Eftychios A Pnevmatikakis, Daniel Soudry, Yuanjun Gao, Timothy A
Machado, Josh Merel, David Pfau, Thomas Reardon, Yu Mu, Clay
Lacefield, Weijian Yang, et al. Simultaneous denoising, deconvolution, and
demixing of calcium imaging data. Neuron, 89(2):285–299, 2016.
P Zhou, SL Resendez, GD Stuber, RE Kass, and L Paninski. Efficient
and accurate extraction of in vivo calcium signals from microendoscope
video data. arXiv preprint arXiv:1605.07266, 2016.
Pengcheng Zhou, Shanna L Resendez, Jose Rodriguez-Romaguera,
Jessica C Jimenez, Shay Q Neufeld, Andrea Giovannucci, Johannes
Friedrich, Eftychios A Pnevmatikakis, Garret D Stuber, Rene Hen,
et al. Efficient and accurate extraction of in vivo calcium signals from
microendoscopic video data. ELife, 7:e28728, 2018.
Noah Apthorpe, Alexander Riordan, Robert Aguilar, Jan Homann,
Yi Gu, David Tank, and H Sebastian Seung. Automatic neuron detection
in calcium imaging data using convolutional networks. In Advances in
Neural Information Processing Systems, pages 3270–3278, 2016.
Alexander Dubbs, James Guevara, and Rafael Yuste. moco: Fast motion
correction for calcium imaging. Frontiers in neuroinformatics, 10:6, 2016.
Raphaël Marée, Loïc Rollus, Benjamin Stévens, Renaud Hoyoux,
Gilles Louppe, Rémy Vandaele, Jean-Michel Begon, Philipp Kainz, Pierre
Geurts, and Louis Wehenkel. Collaborative analysis of multi-gigapixel
imaging data using cytomine. Bioinformatics, 32(9):1395–1401, 2016.
Ali I Mohammed, Howard J Gritton, Hua-an Tseng, Mark E Bucklin,
Zhaojie Yao, and Xue Han. An integrative approach for analyzing hundreds
of neurons in task performing mice using wide-field calcium imaging.
Scientific reports, 6(1):20986, 2016.
B. Ahanonu, L. J. Kitch, T. H. Kim, M. C. Larkin, E. O. Hamel,
J. Lecoq, D. E. Aldarondo, and M. J. Schnitzer. Maximum likelihood and
machine learning based methods for automated cell sorting of large-scale
neural calcium imaging data. Society for Neuroscience, 2018. URL
https://abstractsonline.com/pp8/#!/4649/presentation/41917.
B. Ahanonu, L. J. Kitch, T. H. Kim, M. C. Larkin,
E. O. Hamel, J. Lecoq, and M. J. Schnitzer. Maximum
likelihood based cell sorting of large-scale neural calcium
imaging data. Society for Neuroscience, 2017. URL
http://www.abstractsonline.com/pp8/index.html#!/4376/presentation/18520.
Biafra Owowonta Ahanonu. Neural Ensemble Dynamics in Behaving
Animals: Computational Approaches and Applications in Amygdala and
Striatum. Stanford University, 2018.
Jinghao Lu, Chunyuan Li, and Fan Wang. Seeds cleansing cnmf for
spatiotemporal neural signals extraction of miniscope imaging data. arXiv
preprint arXiv:1704.00793, 2017.
Johannes Friedrich, Pengcheng Zhou, and Liam Paninski. Fast online
deconvolution of calcium imaging data. PLoS computational biology, 13
(3):e1005423, 2017.
Stephanie Reynolds, Therese Abrahamsson, Renaud Schuck, P Jesper
Sjöström, Simon R Schultz, and Pier Luigi Dragotti. Able: An
activity-based level set segmentation algorithm for two-photon calcium
imaging data. eNeuro, pages ENEURO–0012, 2017.
Ashley Petersen, Noah Simon, and Daniela Witten. SCALPEL:
Extracting Neurons from Calcium Imaging Data. ArXiv e-prints, art.
arXiv:1703.06946, March 2017.
Quico Spaen, Dorit S Hochbaum, and Roberto Asín-Achá. Hnccorr:
A novel combinatorial approach for cell identification in calcium-imaging
movies. arXiv preprint arXiv:1703.01999, 2017.
Andrea Giovannucci, Johannes Friedrich, Matt Kaufman, Anne
Churchland, Dmitri Chklovskii, Liam Paninski, and Eftychios A
Pnevmatikakis. Onacid: Online analysis of calcium imaging data in real
time. In Advances in Neural Information Processing Systems, pages
2381–2391, 2017.
Hakan Inan, Murat A Erdogdu, and Mark Schnitzer. Robust estimation
of neural signals in calcium imaging. In Advances in Neural Information
Processing Systems, pages 2901–2910, 2017.
JG Orlandi,
S Fernández-García, A Comella-Bolla, M Masana, G García-Díaz
Barriga, M Yaghoubi, A Kipp, JM Canals, MA Colicos, J Davidsen,
et al. Netcal: An interactive platform for large-scale, network and
population dynamics analysis of calcium imaging recordings, zenodo
(2017).
Eftychios A Pnevmatikakis and Andrea Giovannucci. Normcorre: An
online algorithm for piecewise rigid motion correction of calcium imaging
data. Journal of neuroscience methods, 291:83–94, 2017.
Liron Sheintuch, Alon Rubin, Noa Brande-Eilat, Nitzan Geva, Noa Sadeh,
Or Pinchasof, and Yaniv Ziv. Tracking the same neurons across multiple
days in ca2+ imaging data. Cell reports, 21(4):1102–1115, 2017.
Jiangheng Guan, Jingcheng Li, Shanshan Liang, Ruijie Li, Xingyi Li,
Xiaozhe Shi, Ciyu Huang, Jianxiong Zhang, Junxia Pan, Hongbo Jia,
et al. Neuroseg: automated cell detection and segmentation for in vivo
two-photon ca 2+ imaging data. Brain Structure and Function, 223(1):
519–533, 2018.
Takashi Takekawa, Hirotaka Asai, Noriaki Ohkawa, Masanori Nomoto,
Reiko Okubo-Suzuki, Khaled Ghandour, Masaaki Sato, Yasunori Hayashi,
Kaoru Inokuchi, and Tomoki Fukai. Automatic sorting system for large
calcium imaging data. bioRxiv, page 215145, 2017.
Sebastián A Romano, Verónica Pérez-Schuster, Adrien Jouary,
Jonathan Boulanger-Weill, Alessia Candeo, Thomas Pietri, and Germán
Sumbre. An integrated calcium imaging processing toolbox for the analysis
of neuronal population dynamics. PLoS computational biology, 13(6):
e1005526, 2017a.
Martin Rueckl, Stephen C Lenzi, Laura Moreno-Velasquez, Daniel
Parthier, Dietmar Schmitz, Sten Ruediger, and Friedrich W Johenning.
Samuroi, a python-based software tool for visualization and analysis of
dynamic time series imaging at multiple spatial scales. Frontiers in
neuroinformatics, 11:44, 2017.
Alexander Fillbrunn, Christian Dietz, Julianus Pfeuffer, René Rahn,
Gregory A Landrum, and Michael R Berthold. Knime for reproducible
cross-domain analysis of life science data. Journal of biotechnology, 261:
149–156, 2017.
Aleksander Klibisz, Derek Rose, Matthew Eicholtz, Jay Blundon, and
Stanislav Zakharenko. Fast, simple calcium imaging segmentation with
fully convolutional networks. In International Workshop on Deep Learning
in Medical Image Analysis, pages 285–293. Springer, 2017.
Sander W Keemink, Scott C Lowe, Janelle MP Pakan, Evelyn Dylda,
Mark CW Van Rossum, and Nathalie L Rochefort. Fissa: A neuropil
decontamination toolbox for calcium imaging signals. Scientific reports, 8
(1):1–12, 2018.
Gal Mishne, Ronald R Coifman, Maria Lavzin, and Jackie Schiller.
Automated cellular structure extraction in biological images with
applications to calcium imaging data. bioRxiv, page 313981, 2018.
E Kelly Buchanan, Ian Kinsella, Ding Zhou, Rong Zhu, Pengcheng Zhou,
Felipe Gerhard, John Ferrante, Ying Ma, Sharon Kim, Mohammed Shaik,
et al. Penalized matrix decomposition for denoising, compression, and
improved demixing of functional imaging data. bioRxiv, page 334706, 2018.
Jinghao Lu, Chunyuan Li, Jonnathan Singh-Alvarado, Zhe Charles
Zhou, Flavio Fröhlich, Richard Mooney, and Fan Wang. MIN1PIPE: A
Miniscope 1-Photon-Based Calcium Imaging Signal Extraction Pipeline.
Cell Reports, 23(12):3673–3684, 2018. ISSN 2211-1247.
Andrea Giovannucci, Johannes Friedrich, Pat Gunn, Jeremie Kalfon,
Sue Ann Koay, Jiannis Taxidis, Farzaneh Najafi, Jeffrey L Gauthier,
Pengcheng Zhou, and David W Tank. CaImAn: An open source tool for
scalable Calcium Imaging data Analysis. bioRxiv, page 339564, 2018.
Jeffrey L Gauthier, Sue Ann Koay, Edward H Nieh, David W Tank,
Jonathan W Pillow, and Adam S Charles. Detecting and correcting false
transients in calcium imaging. bioRxiv, page 473470, 2018.
Simon P Shen, Hua-an Tseng, Kyle R Hansen, Ruofan Wu, Howard J
Gritton, Jennie Si, and Xue Han. Automatic cell segmentation by adaptive
thresholding (acsat) for large-scale calcium imaging datasets. eneuro, 5(5),
2018.
Akinori Mitani and Takaki Komiyama. Real-time processing of
two-photon calcium imaging data including lateral motion artifact
correction. Frontiers in neuroinformatics, 12:98, 2018.
Gregory Corder, Biafra Ahanonu, Benjamin F Grewe, Dong Wang,
Mark J Schnitzer, and Grégory Scherrer. An amygdalar neural ensemble
that encodes the unpleasantness of pain. Science, 363(6424):276–281, 2019.
Biafra Ahanonu and Gregory Corder. Recording pain-related brain
activity in behaving animals using calcium imaging calcium imaging and
miniature microscopes. In Contemporary Approaches to the Study of Pain:
From Molecules to Neural Networks, pages 217–276. Springer, 2022.
Adam S Charles, Alex Song, Jeffrey L Gauthier, Jonathan W Pillow,
and David W Tank. Neural anatomy and optical microscopy (naomi)
simulation for evaluating calcium imaging methods. bioRxiv, page 726174,
2019.
FDW Radstake, EAL Raaijmakers, R Luttge, Svitlana Zinger, and
Jean-Philippe Frimat. Calima: The semi-automated open-source calcium
imaging analyzer. Computer methods and programs in biomedicine, 179:
104991, 2019.
Somayyeh Soltanian-Zadeh, Kaan Sahingur, Sarah Blau, Yiyang Gong,
and Sina Farsiu. Fast and robust active neuron segmentation in two-photon
calcium imaging using spatiotemporal deep learning. Proceedings of the
National Academy of Sciences, 116(17):8554–8563, 2019.
Yizhi Wang, Nicole V DelRosso, Trisha V Vaidyanathan, Michelle K
Cahill, Michael E Reitman, Silvia Pittolo, Xuelong Mi, Guoqiang Yu,
and Kira E Poskanzer. Accurate quantification of astrocyte and
neurotransmitter fluorescence dynamics for single-cell and population-level
physiology. Nature Neuroscience, 22(11):1936–1944, 2019.
Andrea Giovannucci, Johannes Friedrich, Pat Gunn, Jeremie Kalfon,
Brandon L Brown, Sue Ann Koay, Jiannis Taxidis, Farzaneh Najafi,
Jeffrey L Gauthier, Pengcheng Zhou, et al. Caiman an open source tool
for scalable calcium imaging data analysis. Elife, 8:e38173, 2019.
Gal Mishne and Adam S Charles. Learning spatially-correlated
temporal dictionaries for calcium imaging. In ICASSP 2019-2019 IEEE
International Conference on Acoustics, Speech and Signal Processing
(ICASSP), pages 1065–1069. IEEE, 2019.
Noah Dolev, Lior Pinkus, and Michal Rivlin-Etzion. Segment2p:
Parameter-free automated segmentation of cellular fluorescent signals.
BioRxiv, page 832188, 2019.
Zhe Chen, Hugh T Blair, and Jason Cong. Lanmc: Lstm-assisted
non-rigid motion correction on fpga for calcium image stabilization.
In Proceedings of the 2019 ACM/SIGDA International Symposium on
Field-Programmable Gate Arrays, pages 104–109, 2019.
Ryohei Shibue and Fumiyasu Komaki. Deconvolution of calcium imaging
data using marked point processes. PLoS computational biology, 16(3):
e1007650, 2020.
Shreya Saxena, Ian Kinsella, Simon
Musall, Sharon H Kim, Jozsef Meszaros, David N Thibodeaux, Carla
Kim, John Cunningham, Elizabeth MC Hillman, Anne Churchland, et al.
Localized semi-nonnegative matrix factorization (locanmf) of widefield
calcium imaging data. PLOS Computational Biology, 16(4):e1007791,
2020.
Daniel A Cantu, Bo Wang, Michael W Gongwer, Cynthia X He,
Anubhuti Goel, Anand Suresh, Nazim Kourdougli, Erica D Arroyo,
William Zeiger, and Carlos Portera-Cailliau. Ezcalcium: Open source
toolbox for analysis of calcium imaging data. bioRxiv, 2020.
Johannes Friedrich, Andrea Giovannucci, and
Eftychios A Pnevmatikakis. Online analysis of microendoscopic 1-photon
calcium imaging data streams. bioRxiv, 2020.
Lina M Tran, Andrew J Mocle, Adam I Ramsaran, Alex D Jacob,
Paul W Frankland, and Sheena A Josselyn. Automated curation
of cnmf-e-extracted roi spatial footprints and calcium traces using
open-source automl tools. bioRxiv, 2020a.
Lina M Tran, Andrew J Mocle, Adam I Ramsaran, Alexander D
Jacob, Paul W Frankland, and Sheena A Josselyn. Automated curation
of cnmf-e-extracted roi spatial footprints and calcium traces using
open-source automl tools. Frontiers in Neural Circuits, 14:42, 2020b.
Jerome Lecoq, Michael Oliver, Joshua H Siegle, Natalia Orlova, and
Christof Koch. Removing independent noise in systems neuroscience data
using deepinterpolation. bioRxiv, 2020.
Ulysse Rubens, Romain Mormont, Lassi Paavolainen, Volker Bäcker,
Benjamin Pavie, Leandro A Scholz, Gino Michiels, Martin Maška,
Devrim Ünay, Graeme Ball, et al. Biaflows: A collaborative framework
to reproducibly deploy and benchmark bioimage analysis workflows.
Patterns, 1(3):100040, 2020.
Ryan M Cassidy, Alexis G Bavencoffe, Elia R Lopez, Sai S Cheruvu,
Edgar T Walters, Rosa A Uribe, Anne Marie Krachler, and Max A
Odem. Frequency-independent biological signal identification (fibsi): A
free program that simplifies intensive analysis of non-stationary time series
data. bioRxiv, 2020.
Sascha RA Alles, Max A Odem, Van B Lu, Ryan M Cassidy, and
Peter A Smith. Chronic bdnf simultaneously inhibits and unmasks
superficial dorsal horn neuronal activity. Scientific reports, 11(1):1–14,
2021.
Elke Kirschbaum, Alberto Bailoni, and Fred A Hamprecht. Disco: deep
learning, instance segmentation, and correlations for cell segmentation in
calcium imaging. In Medical Image Computing and Computer Assisted
Intervention–MICCAI 2020: 23rd International Conference, Lima, Peru,
October 4–8, 2020, Proceedings, Part V 23, pages 151–162. Springer, 2020.
Julien Denis, Robin F Dard, Eleonora Quiroli, Rosa Cossart, and
Michel A Picardo. Deepcinac: a deep-learning-based python toolbox for
inferring calcium imaging neuronal activity based on movie visualization.
eneuro, 7(4), 2020.
Yaesop Lee, Jing Xie, Eungjoo Lee, Srijesh Sudarsanan, Da-Ting Lin,
Rong Chen, and Shuvra S Bhattacharyya. Real-time neuron detection and
neural signal extraction platform for miniature calcium imaging. Frontiers
in Computational Neuroscience, 14:43, 2020.
Chaozhen Tan, Yue Guan, Zhao Feng, Hong Ni, Zoutao Zhang, Zhiguang
Wang, Xiangning Li, Jing Yuan, Hui Gong, Qingming Luo, et al.
Deepbrainseg: Automated brain region segmentation for micro-optical
images with a convolutional neural network. Frontiers in neuroscience, 14:
179, 2020.
Victoria A Griffiths, Antoine M Valera, Joanna YN Lau, Hana
Roš, Thomas J Younts, Bóris Marin, Chiara Baragli, Diccon Coyle,
Geoffrey J Evans, George Konstantinou, et al. Real-time 3d movement
correction for two-photon imaging in behaving animals. Nature methods,
17(7):741–748, 2020.
Carsen Stringer, Tim Wang, Michalis Michaelos, and Marius Pachitariu.
Cellpose: a generalist algorithm for cellular segmentation. Nature Methods,
18(1):100–106, 2021.
Alexander Song, Jeff L Gauthier, Jonathan W Pillow, David W Tank,
and Adam S Charles. Neural anatomy and optical microscopy (naomi)
simulation for evaluating calcium imaging methods. Journal of
Neuroscience Methods, 358:109173, 2021.
Johannes
Friedrich, Andrea Giovannucci, and Eftychios A Pnevmatikakis. Online
analysis of microendoscopic 1-photon calcium imaging data streams. PLoS
computational biology, 17(1):e1008565, 2021.
Hakan Inan, Claudia Schmuckermair, Tugce Tasci, Biafra Ahanonu,
Oscar Hernandez, Jérôme Lecoq, Fatih Dinç, Mark J Wagner, Murat
Erdogdu, and Mark J Schnitzer. Fast and statistically robust cell
extraction from large-scale neural calcium imaging datasets. bioRxiv, 2021.
Zhe Dong, William Mau, Yu Susie Feng, Zachary T Pennington,
Lingxuan Chen, Yosif Zaki, Kanaka Rajan, Tristan Shuman, Daniel
Aharoni, and Denise J Cai. Minian: An open-source miniscope analysis
pipeline. bioRxiv, 2021.
Kushal Kolar, Daniel Dondorp, Jordi Cornelis Zwiggelaar, Jørgen
Høyer, and Marios Chatzigeorgiou. Mesmerize: a dynamically adaptable
user-friendly analysis platform for 2d & 3d calcium imaging data. bioRxiv,
page 840488, 2021.
Jérôme Lecoq, Michael Oliver, Joshua H Siegle, Natalia Orlova, Peter
Ledochowitsch, and Christof Koch. Removing independent noise in
systems neuroscience data using deepinterpolation. Nature Methods, pages
1–8, 2021.
Seungjae Han, Eun-Seo Cho, Inkyu Park, Kijung Shin, and Young-Gyu
Yoon. Efficient neural network approximation of robust pca for automated
analysis of calcium imaging data. In International Conference on Medical
Image Computing and Computer-Assisted Intervention, pages 595–604.
Springer, 2021.
Madhavi Tippani, Elizabeth A Pattie, Brittany A Davis, Claudia V
Nguyen, Yanhong Wang, Srinidhi Rao Sripathy, Brady J Maher, Keri
Martinowich, Andrew E Jaffe, and Stephanie Cerceo Page. Capture:
Calcium peak toolbox for analysis of in vitro calcium imaging data.
bioRxiv, 2021.
Peter Rupprecht, Stefano Carta, Adrian Hoffmann, Mayumi Echizen,
Antonin Blot, Alex C Kwan, Yang Dan, Sonja B Hofer, Kazuo Kitamura,
Fritjof Helmchen, et al. A database and deep learning toolbox for
noise-optimized, generalized spike inference from calcium imaging. Nature
Neuroscience, 24(9):1324–1337, 2021.
Changjia Cai, Johannes Friedrich, Amrita Singh, M Hossein Eybposh,
Eftychios A Pnevmatikakis, Kaspar Podgorski, and Andrea Giovannucci.
Volpy: automated and scalable analysis pipelines for voltage imaging
datasets. PLoS computational biology, 17(4):e1008806, 2021.
Xinyang Li, Guoxun Zhang, Jiamin Wu, Yuanlong Zhang, Zhifeng Zhao,
Xing Lin, Hui Qiao, Hao Xie, Haoqian Wang, Lu Fang, et al. Reinforcing
neuron extraction and spike inference in calcium imaging using deep
self-supervised denoising. Nature Methods, pages 1–6, 2021.
Leander
de Kraker, Koen Seignette, Premnath Thamizharasu, Bastijn JG van den
Boom, Ildefonso Ferreira Pica, Ingo Willuhn, Christiaan N Levelt, and
Chris van der Togt. Specseg: cross spectral power-based segmentation of
neurons and neurites in chronic calcium imaging datasets. bioRxiv, pages
2020–10, 2021.
Andrea Giovannucci, Changjia Cai, Cynthia Dong, Marton Rozsa, and
Eftychios Pnevmatikakis. Fiola: An accelerated pipeline for fluorescence
imaging online analysis. 2021.
Ryoma Hattori and Takaki Komiyama. Patchwarp:
Corrections of non-uniform image distortions in two-photon
calcium imaging data by patchwork affine transformations.
bioRxiv, 2021. doi: 10.1101/2021.11.10.468164. URL
https://www.biorxiv.org/content/early/2021/11/13/2021.11.10.468164.
Xiaohui Zhang, Eric C Landsness, Wei Chen, Hanyang Miao, Michelle
Tang, Lindsey M Brier, Joseph P Culver, Jin-Moo Lee, and Mark A
Anastasio. Automated sleep state classification of wide-field calcium
imaging data via multiplex visibility graphs and deep learning. Journal of
Neuroscience Methods, page 109421, 2021.
Philipp Flotho, Shinobu Nomura, Bernd Kuhn, and Daniel J Strauss.
Software for non-parametric image registration of 2-photon imaging data.
Journal of Biophotonics, 15(8):e202100330, 2022.
Yijun Bao, Somayyeh Soltanian-Zadeh, Sina Farsiu, and Yiyang Gong.
Segmentation of neurons from fluorescence calcium recordings beyond real
time. Nature machine intelligence, 3(7):590–600, 2021.
Masaki Taniguchi, Taro Tezuka, Pablo Vergara, Sakthivel Srinivasan,
Takuma Hosokawa, Yoan Chérasse, Toshie Naoi, Takeshi Sakurai,
and Masanori Sakaguchi. Open-source software for real-time calcium
imaging and synchronized neuron firing detection. In 2021 43rd Annual
International Conference of the IEEE Engineering in Medicine & Biology
Society (EMBC), pages 2997–3003. IEEE, 2021.
Brian R Mullen, Sydney C Weiser, Desiderio Ascencio, and James B
Ackman. Automated classification of signal sources in mesoscale calcium
imaging. bioRxiv, pages 2021–02, 2021.
Yina Wang, Henry Pinkard, Emaad Khwaja, Shuqin Zhou, Laura
Waller, and Bo Huang. Image denoising for fluorescence microscopy by
self-supervised transfer learning. bioRxiv, pages 2021–02, 2021.
Thibault Lagache, Alison Hanson, Jesús E Pérez-Ortega, Adrienne
Fairhall, and Rafael Yuste. Tracking calcium dynamics from individual
neurons in behaving animals. PLOS Computational Biology, 17(10):
e1009432, 2021.
Adam S Charles, Nathan Cermak, Rifqi O Affan, Benjamin B Scott,
Jackie Schiller, and Gal Mishne. Graft: graph filtered temporal dictionary
learning for functional neural imaging. IEEE Transactions on Image
Processing, 31:3509–3524, 2022.
Madhavi Tippani, Elizabeth A Pattie, Brittany A Davis, Claudia V
Nguyen, Yanhong Wang, Srinidhi Rao Sripathy, Brady J Maher, Keri
Martinowich, Andrew E Jaffe, and Stephanie Cerceo Page. Capture:
Calcium peaktoolbox for analysis of in vitro calcium imaging data. BMC
neuroscience, 23(1):71, 2022.
Leander de Kraker, Koen Seignette,
Premnath Thamizharasu, Bastijn JG van den Boom, Ildefonso Ferreira
Pica, Ingo Willuhn, Christiaan N Levelt, and Chris van der Togt. Specseg
is a versatile toolbox that segments neurons and neurites in chronic calcium
imaging datasets based on low-frequency cross-spectral power. Cell reports
methods, 2(10), 2022.
Luca Sità, Marco Brondi, Pedro Lagomarsino de Leon Roig, Sebastiano
Curreli, Mariangela Panniello, Dania Vecchia, and Tommaso Fellin. A
deep-learning approach for online cell identification and trace extraction in
functional two-photon calcium imaging. Nature Communications, 13(1):
1529, 2022.
Honghua Guan, Dawei Li, Hyeon-cheol Park, Ang Li, Yuanlei Yue,
Yung-Tian A Gau, Ming-Jun Li, Dwight E Bergles, Hui Lu, and Xingde
Li. Deep-learning two-photon fiberscopy for video-rate brain imaging in
freely-behaving mice. Nature communications, 13(1):1534, 2022.
Sharif Amit Kamran, Khondker Fariha Hossain, Hussein Moghnieh,
Sarah Riar, Allison Bartlett, Alireza Tavakkoli, Kenton M Sanders, and
Salah A Baker. New open-source software for subcellular segmentation
and analysis of spatiotemporal fluorescence signals using deep learning.
Iscience, 25(5), 2022.
Xinyang Li, Yixin Li, Yiliang Zhou, Jiamin Wu, Zhifeng Zhao, Jiaqi
Fan, Fei Deng, Zhaofa Wu, Guihua Xiao, Jing He, et al. Real-time
denoising enables high-sensitivity fluorescence time-lapse imaging beyond
the shot-noise limit. Nature Biotechnology, 41(2):282–292, 2023a.
Jeffrey L Gauthier, Sue Ann Koay, Edward H Nieh, David W Tank,
Jonathan W Pillow, and Adam S Charles. Detecting and correcting false
transients in calcium imaging. Nature Methods, 19(4):470–478, 2022.
Andres Flores-Valle and Johannes D Seelig. Axial motion estimation
and correction for simultaneous multi-plane two-photon calcium imaging.
Biomedical Optics Express, 13(4):2035–2049, 2022.
Weiyi Liu, Junxia Pan, Yuanxu Xu, Meng Wang, Hongbo Jia, Kuan
Zhang, Xiaowei Chen, Xingyi Li, and Xiang Liao. Fast and accurate motion
correction for two-photon ca2+ imaging in behaving mice. Frontiers in
Neuroinformatics, 16:851188, 2022.
Oliver Rübel, Andrew Tritt, Ryan Ly, Benjamin K Dichter, Satrajit
Ghosh, Lawrence Niu, Pamela Baker, Ivan Soltesz, Lydia Ng, Karel
Svoboda, et al. The neurodata without borders ecosystem for
neurophysiological data science. Elife, 11:e78362, 2022.
Zhe Chen, Garrett J Blair, Changliang Guo, Jim Zhou, Juan-Luis
Romero-Sosa, Alicia Izquierdo, Peyman Golshani, Jason Cong, Daniel
Aharoni, and Hugh T Blair. A hardware system for real-time decoding of
in vivo calcium imaging data. Elife, 12:e78344, 2023.
Yan Zhang, Márton Rózsa, Yajie Liang, Daniel Bushey, Ziqiang
Wei, Jihong Zheng, Daniel Reep, Gerard Joey Broussard, Arthur Tsang,
Getahun Tsegaye, et al. Fast and sensitive gcamp calcium indicators for
imaging neural populations. Nature, 615(7954):884–891, 2023a.
Zhehao Xu, Yukun Wu, Jiangheng Guan, Shanshan Liang, Junxia Pan,
Meng Wang, Qianshuo Hu, Hongbo Jia, Xiaowei Chen, and Xiang Liao.
Neuroseg-ii: A deep learning approach for generalized neuron segmentation
in two-photon ca2+ imaging. Frontiers in Cellular Neuroscience, 17:
1127847, 2023.
Pablo Vergara, Yuteng Wang, Sakthivel Srinivasan, Yoan Cherasse,
Toshie Naoi, Yuki Sugaya, Takeshi Sakurai, Masanobu Kano, and Masanori
Sakaguchi. The caliali tool for long-term tracking of neuronal population
dynamics in calcium imaging. bioRxiv, pages 2023–05, 2023.
Yuanlong Zhang, Guoxun Zhang, Xiaofei Han, Jiamin Wu, Ziwei Li,
Xinyang Li, Guihua Xiao, Hao Xie, Lu Fang, and Qionghai Dai. Rapid
detection of neurons in widefield calcium imaging datasets after training
with synthetic data. Nature Methods, 20(5):747–754, 2023b.
Jacopo Bonato, Sebastiano Curreli, Sara Romanzi, Stefano Panzeri, and
Tommaso Fellin. Astra: a deep learning algorithm for fast semantic
segmentation of large-scale astrocytic networks. bioRxiv, pages 2023–05,
2023.
Xinyang Li, Xiaowan Hu, Xingye Chen, Jiaqi Fan, Zhifeng Zhao, Jiamin
Wu, Haoqian Wang, and Qionghai Dai. Spatial redundancy transformer
for self-supervised fluorescence image denoising. bioRxiv, pages 2023–06,
2023b.
Junmo Cho, Seungjae Han, Eun-Seo Cho, Kijung Shin, and Young-Gyu
Yoon. Robust and efficient alignment of calcium imaging data through
simultaneous low rank and sparse decomposition. In Proceedings of the
IEEE/CVF Winter Conference on Applications of Computer Vision, pages
1939–1948, 2023.
Biafra Ahanonu, Andrew Crowther, Artur Kania, Mariela Rosa Casillas,
and Allan Basbaum. Long-term optical imaging of the spinal cord in awake,
behaving animals. bioRxiv, pages 2023–05, 2023.
Sebastián A Romano, Verónica Pérez-Schuster, Adrien Jouary,
Alessia Candeo, Jonathan Boulanger-Weill, and Germán Sumbre. A
computational toolbox and step-by-step tutorial for the analysis of
neuronal population dynamics in calcium imaging data. bioRxiv, page
103879, 2017b.
Kunal K Ghosh, Laurie D Burns, Eric D Cocker, Axel Nimmerjahn,
Yaniv Ziv, Abbas El Gamal, and Mark J Schnitzer. Miniaturized
integration of a fluorescence microscope. Nature Methods, 8(10):871–878,
2011.
Yaniv Ziv, Laurie D Burns, Eric D Cocker, Elizabeth O Hamel,
Kunal K Ghosh, Lacey J Kitch, Abbas El Gamal, and Mark J Schnitzer.
Long-term dynamics of ca1 hippocampal place codes. Nature neuroscience,
16(3):264–266, 2013.
Elizabeth J.O. Hamel, Benjamin F. Grewe, Jones G. Parker, and
Mark J. Schnitzer. Cellular Level Brain Imaging in Behaving
Mammals: An Engineering Approach. Neuron, 86(1):140–159,
2015. ISSN 0896-6273. doi: 10.1016/j.neuron.2015.03.055. URL
http://www.sciencedirect.com/science/article/pii/S0896627315002792{\%}5Cnhttp://linkinghub.elsevier.com/retrieve/pii/S0896627315002792.
R Madelaine Paredes, Julie C Etzler, Lora Talley Watts, Wei Zheng,
and James D Lechleiter. Chemical calcium indicators. Methods, 46(3):
143–151, 2008.
Oded Tour, Stephen R Adams, Rex A Kerr, Rene M Meijer, Terrence J
Sejnowski, Richard W Tsien, and Roger Y Tsien. Calcium green flash as
a genetically targeted small-molecule calcium indicator. Nature chemical
biology, 3(7):423, 2007.
Tsai-Wen Chen, Trevor J Wardill, Yi Sun, Stefan R Pulver, Sabine L
Renninger, Amy Baohan, Eric R Schreiter, Rex a Kerr, Michael B Orger,
Vivek Jayaraman, Loren L Looger, Karel Svoboda, and Douglas S Kim.
Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature,
499(7458):295–300, July 2013. ISSN 1476-4687. doi: 10.1038/nature12354.
URL http://www.ncbi.nlm.nih.gov/pubmed/23868258.
Hod Dana, Yi Sun, Boaz Mohar, Brad Hulse, Jeremy P Hasseman,
Getahun Tsegaye, Arthur Tsang, Allan Wong, Ronak Patel, and John J
Macklin. High-performance GFP-based calcium indicators for imaging
activity in neuronal populations and microcompartments. bioRxiv, page
434589, 2018.
Marco Mank, Alexandre Ferrao Santos, Stephan Direnberger, Thomas D
Mrsic-Flogel, Sonja B Hofer, Valentin Stein, Thomas Hendel, Dierk F
Reiff, Christiaan Levelt, Alexander Borst, et al. A genetically encoded
calcium indicator for chronic in vivo two-photon imaging. Nature methods,
5(9):805, 2008.
Gerard Joey Broussard, Yajie Liang, Marina Fridman, Elizabeth K
Unger, Guanghan Meng, Xian Xiao, Na Ji, Leopoldo Petreanu, and Lin
Tian. In vivo measurement of afferent activity with axon-specific calcium
imaging. Technical report, Nature Publishing Group, 2018.
Elena Dreosti, Benjamin Odermatt, Mario M Dorostkar, and Leon
Lagnado. A genetically encoded reporter of synaptic activity in vivo.
Nature methods, 6(12):883, 2009.
Tony Hyun Kim, Yanping Zhang, Jérôme Lecoq, Juergen C Jung,
Jane Li, Hongkui Zeng, Cristopher M Niell, and Mark J Schnitzer.
Long-Term Optical Access to an Estimated One Million Neurons in the
Live Mouse Cortex. Cell Reports, 17(12):3385–3394, 2016. ISSN 2211-1247.
Saskia E J de Vries, Jerome Lecoq, Michael A Buice, Peter A
Groblewski, Gabriel K Ocker, Michael Oliver, David Feng, Nicholas Cain,
Peter Ledochowitsch, and Daniel Millman. A large-scale, standardized
physiological survey reveals higher order coding throughout the mouse
visual cortex. bioRxiv, page 359513, 2018.
Bjorn L Millard, Mario Niepel, Michael P Menden, Jeremy L Muhlich,
and Peter K Sorger. Adaptive informatics for multifactorial and
high-content biological data. Nature methods, 8(6):487–492, 2011.
Alfie Kohn. Why incentive plans cannot work. Harvard business review,
71(5), 1993a.
Alfie Kohn. For best results, forget the bonus. New York Times, 17:F11,
1993b.
Uri Gneezy and Aldo Rustichini. Pay enough or don’t pay at all.
Quarterly journal of economics, pages 791–810, 2000.
Gaetan de Lavilléon, Marie Masako Lacroix, Laure Rondi-Reig, and
Karim Benchenane. Explicit memory creation during sleep demonstrates a
causal role of place cells in navigation. Nature neuroscience, 18(4):493–495,
2015.
Patricia H Janak and Kay M Tye. From circuits to behaviour in the
amygdala. Nature, 517(7534):284–292, 2015.
Matthew B Bouchard, Venkatakaushik Voleti, César S Mendes, Clay
Lacefield, Wesley B Grueber, Richard S Mann, Randy M Bruno, and
Elizabeth MC Hillman. Swept confocally-aligned planar excitation (scape)
microscopy for high-speed volumetric imaging of behaving organisms.
Nature Photonics, 2015.
Elizabeth MC Hillman, Anna Devor, Andrew K Dunn, and David A
Boas. Laminar optical tomography: high-resolution 3d functional imaging
of superficial tissues. In Medical Imaging, pages 61431M–61431M.
International Society for Optics and Photonics, 2006.
Michaela Mickoleit, Benjamin Schmid, Michael Weber, Florian O
Fahrbach, Sonja Hombach, Sven Reischauer, and Jan Huisken.
High-resolution reconstruction of the beating zebrafish heart. Nature
methods, 2014.
Volodymyr Mnih, Koray Kavukcuoglu, David Silver, Andrei A Rusu, Joel
Veness, Marc G Bellemare, Alex Graves, Martin Riedmiller, Andreas K
Fidjeland, Georg Ostrovski, et al. Human-level control through deep
reinforcement learning. Nature, 518(7540):529–533, 2015.
Jeff R Jones, Michael C Tackenberg, and Douglas G McMahon.
Manipulating circadian clock neuron firing rate resets molecular circadian
rhythms and behavior. Nature neuroscience, 18(3):373–375, 2015.
Benjamin Rusak and Gerard Groos. Suprachiasmatic stimulation phase
shifts rodent circadian rhythms. Science, 215(4538):1407–1409, 1982.
Paula E Stephan and Sharon G Levin. Age and the nobel prize revisited.
Scientometrics, 28(3):387–399, 1993.
Matthew E Falagas, Vrettos Ierodiakonou, and Vangelis G Alexiou. At
what age do biomedical scientists do their best work? The FASEB Journal,
22(12):4067–4070, 2008.
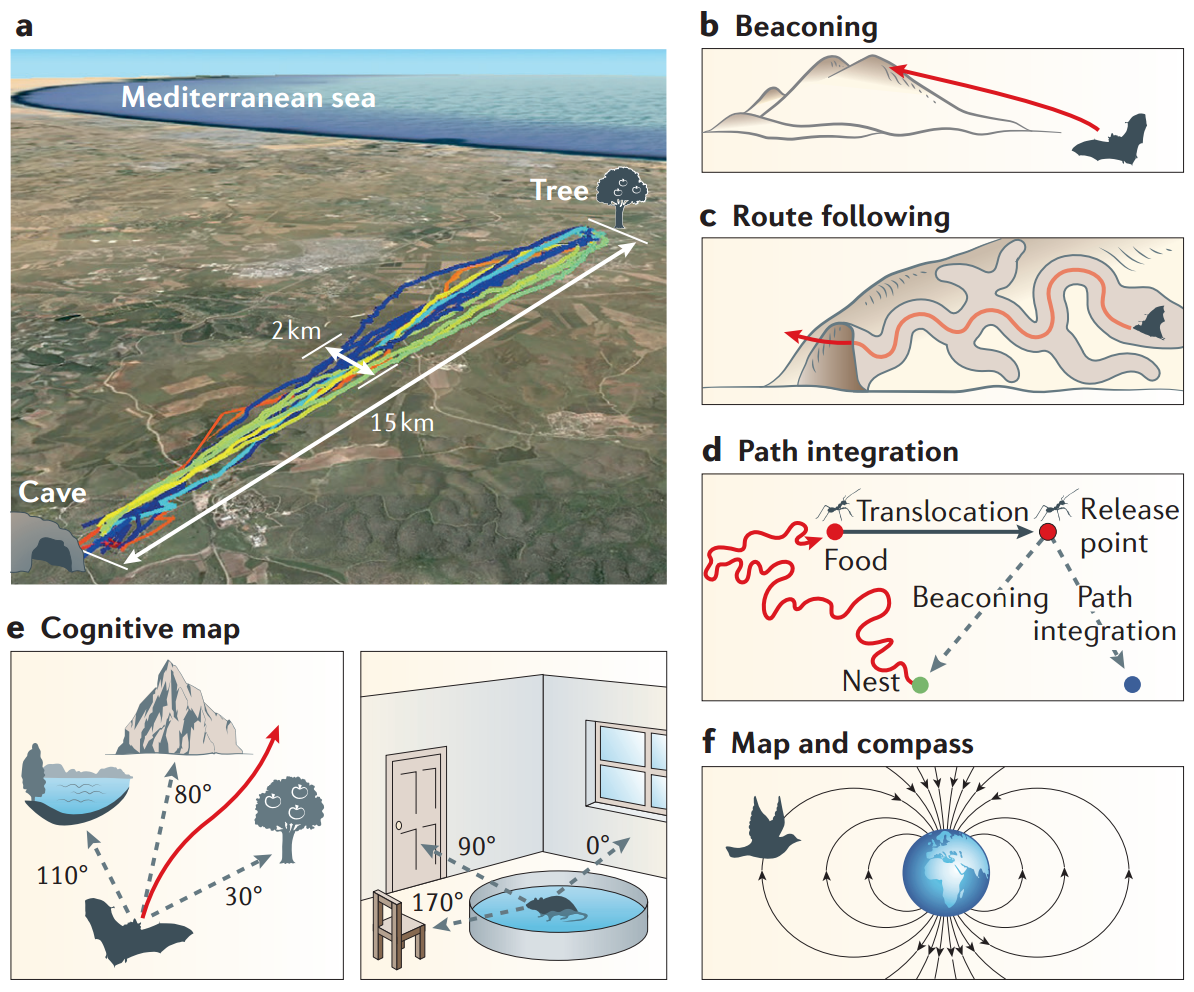
Maya Geva-Sagiv, Liora Las, Yossi Yovel, and Nachum Ulanovsky. Spatial
cognition in bats and rats: from sensory acquisition to multiscale maps and
navigation. Nature Reviews Neuroscience, 16(2):94–108, 2015.
Rajesh PN Rao, Andrea Stocco, Matthew Bryan, Devapratim Sarma,
Tiffany M Youngquist, Joseph Wu, and Chantel S Prat. A direct
brain-to-brain interface in humans. PloS one, 9(11):e111332, 2014.
Frank A Beach. The snark was a boojum. American Psychologist, 5(4):
115, 1950.
Adam M Packer, Lloyd E Russell, Henry WP Dalgleish, and Michael
Häusser. Simultaneous all-optical manipulation and recording of neural
circuit activity with cellular resolution in vivo. Nature methods, 2014.
John Peter Rickgauer, Karl Deisseroth, and David W
Tank. Simultaneous cellular-resolution optical perturbation and imaging
of place cell firing fields. Nature neuroscience, 17(12):1816–1824, 2014.
Adam M Packer, Darcy S Peterka, Jan J Hirtz, Rohit Prakash, Karl
Deisseroth, and Rafael Yuste. Two-photon optogenetics of dendritic spines
and neural circuits. Nature methods, 9(12):1202–1205, 2012.
John Peter Rickgauer and David W Tank. Two-photon excitation of
channelrhodopsin-2 at saturation. Proceedings of the National Academy of
Sciences, 106(35):15025–15030, 2009.
C Bushdid, MO Magnasco, LB Vosshall, and A Keller. Humans can
discriminate more than 1 trillion olfactory stimuli. Science, 343(6177):
1370–1372, 2014.
Markus Meister. Can humans really discriminate 1 trillion odors? arXiv
preprint arXiv:1411.0165, 2014.
Tali Weiss, Kobi Snitz, Adi Yablonka, Rehan M Khan, Danyel
Gafsou, Elad Schneidman, and Noam Sobel. Perceptual convergence
of multi-component mixtures in olfaction implies an olfactory white.
Proceedings of the National Academy of Sciences, 109(49):19959–19964,
2012.
Dashun Wang, Chaoming Song, and Albert-László Barabási.
Quantifying long-term scientific impact. Science, 342(6154):127–132, 2013.
Cahir J O’Kane. Drosophila as a model organism for the study
of neuropsychiatric disorders. In Molecular and Functional Models in
Neuropsychiatry, pages 37–60. Springer, 2011.
Anthony M Zador, Joshua Dubnau, Hassana K Oyibo, Huiqing Zhan,
Gang Cao, and Ian D Peikon. Sequencing the connectome. PLoS biology,
10(10):e1001411, 2012.
Thomas Dean, Biafra Ahanonu, Mainak Chowdhury, Anjali Datta, Andre
Esteva, Daniel Eth, Nobie Redmon, Oleg Rumyantsev, and Ysis Tarter.
On the technology prospects and investment opportunities for scalable
neuroscience. Technical report, Google, 2013a.
Yang Lou, Jun Xia, and Lihong V Wang. Mouse brain imaging
using photoacoustic computed tomography. In SPIE BiOS, pages
894340–894340. International Society for Optics and Photonics, 2014.
Nathan C Klapoetke, Yasunobu Murata, Sung Soo Kim, Stefan R
Pulver, Amanda Birdsey-Benson, Yong Ku Cho, Tania K Morimoto,
Amy S Chuong, Eric J Carpenter, Zhijian Tian, et al. Independent
optical excitation of distinct neural populations. Nature methods, 11(3):
338–346, 2014.
Joseph M Brader, Walter Senn, and Stefano Fusi. Learning real-world
stimuli in a neural network with spike-driven synaptic dynamics. Neural
computation, 19(11):2881–2912, 2007.
Saeed Afshar, Omid Kavehei, André van Schaik, Jonathan Tapson, Stan
Skafidas, and Tara Julia Hamilton. Emergence of competitive control in a
memristor-based neuromorphic circuit. In Neural Networks (IJCNN), The
2012 International Joint Conference on, pages 1–8. IEEE, 2012.
Chi-Sang Poon and Kuan Zhou. Neuromorphic silicon neurons and
large-scale neural networks: challenges and opportunities. Frontiers in
neuroscience, 5, 2011.
A. H. Marblestone, B. M. Zamft, Y. G. Maguire, M. G. Shapiro, T. R.
Cybulski, J. I. Glaser, B. Stranges, R. Kalhor, D. A. Dalrymple, D. Seo,
E. Alon, M. M. Maharbiz, J. Carmena, J. Rabaey, E. S. Boyden, G. M.
Church, and K. P. Kording. Physical Principles for Scalable Neural
Recording. ArXiv e-prints, June 2013.
LT Hall, GCG Beart, EA Thomas, DA Simpson, LP McGuinness,
JH Cole, JH Manton, RE Scholten, F Jelezko, Jörg Wrachtrup, et al.
High spatial and temporal resolution wide-field imaging of neuron activity
using quantum nv-diamond. Scientific reports, 2, 2012.
F Dolde, H Fedder, MW Doherty,
T Nöbauer, F Rempp, G Balasubramanian, T Wolf, F Reinhard, LCL
Hollenberg, F Jelezko, et al. Electric-field sensing using single diamond
spins. Nature Physics, 7(6):459–463, 2011.
Viva R Horowitz, Benjamín J Alemán, David J Christle, Andrew N
Cleland, and David D Awschalom. Electron spin resonance of
nitrogen-vacancy centers in optically trapped nanodiamonds. Proceedings
of the National Academy of Sciences, 109(34):13493–13497, 2012.
Tse-Luen Wee, Yan-Kai Tzeng, Chau-Chung Han, Huan-Cheng Chang,
Wunshain Fann, Jui-Hung Hsu, Kuan-Ming Chen, and Yueh-Chung
Yu. Two-photon excited fluorescence of nitrogen-vacancy centers in
proton-irradiated type ib diamond. The Journal of Physical Chemistry A,
111(38):9379–9386, 2007.
Mijail D Serruya, Nicholas G Hatsopoulos, Liam Paninski, Matthew R
Fellows, and John P Donoghue. Brain-machine interface: Instant neural
control of a movement signal. Nature, 416(6877):141–142, 2002.
Jose M Carmena, Mikhail A Lebedev, Roy E Crist, Joseph E
O’Doherty, David M Santucci, Dragan F Dimitrov, Parag G Patil,
Craig S Henriquez, and Miguel AL Nicolelis. Learning to control a
brain–machine interface for reaching and grasping by primates. PLoS
biology, 1(2):e42, 2003.
Joseph E O’Doherty, Mikhail A Lebedev, Peter J Ifft, Katie Z Zhuang,
Solaiman Shokur, Hannes Bleuler, and Miguel AL Nicolelis. Active tactile
exploration using a brain-machine-brain interface. Nature, 479(7372):
228–231, 2011.
C Ethier, ER Oby, MJ Bauman, and LE Miller. Restoration of
grasp following paralysis through brain-controlled stimulation of muscles.
Nature, 485(7398):368–371, 2012.
Leigh R Hochberg, Daniel Bacher, Beata Jarosiewicz, Nicolas Y Masse,
John D Simeral, Joern Vogel, Sami Haddadin, Jie Liu, Sydney S Cash,
Patrick van der Smagt, et al. Reach and grasp by people with tetraplegia
using a neurally controlled robotic arm. Nature, 485(7398):372–375, 2012.
Ben Engelhard, Nofar Ozeri, Zvi Israel, Hagai Bergman, and Eilon
Vaadia. Inducing gamma oscillations and precise spike synchrony by
operant conditioning via brain-machine interface. Neuron, 77(2):361–375,
2013.
Yijun Wang and Tzyy-Ping Jung. A collaborative brain-computer
interface for improving human performance. PloS one, 6(5):e20422, 2011.
Riccardo Poli, Caterina Cinel, Ana Matran-Fernandez, Francisco
Sepulveda, and Adrian Stoica. Towards cooperative brain-computer
interfaces for space navigation. In Proceedings of the 2013 international
conference on Intelligent user interfaces, pages 149–160. ACM, 2013.
Seung-Schik Yoo, Hyungmin Kim, Emmanuel Filandrianos, Seyed Javid
Taghados, and Shinsuk Park. Non-invasive brain-to-brain interface (bbi):
Establishing functional links between two brains. PloS one, 8(4):e60410,
2013.
Tae-il Kim, Jordan G McCall, Yei Hwan Jung, Xian Huang, Edward R
Siuda, Yuhang Li, Jizhou Song, Young Min Song, Hsuan An Pao,
Rak-Hwan Kim, et al. Injectable, cellular-scale optoelectronics with
applications for wireless optogenetics. Science, 340(6129):211–216, 2013.
Thomas Dean, Biafra Ahanonu, Mainak Chowdhury, Anjali Datta, Andre
Esteva, Daniel Eth, Nobie Redmon, Oleg Rumyantsev, and Ysis Tarter.
On the technology prospects and investment opportunities for scalable
neuroscience. arXiv preprint arXiv:1307.7302, 2013b.
Steve Ramirez, Xu Liu, Pei-Ann Lin, Junghyup Suh, Michele Pignatelli,
Roger L. Redondo, Tomás J. Ryan, and Susumu Tonegawa. Creating a
false memory in the hippocampus. Science, 341(6144):387–391, 2013. doi:
10.1126/science.1239073. URL
http://www.sciencemag.org/content/341/6144/387.abstract.
Xu Liu, Steve Ramirez, Petti T Pang, Corey B Puryear, Arvind
Govindarajan, Karl Deisseroth, and Susumu Tonegawa. Optogenetic
stimulation of a hippocampal engram activates fear memory recall. Nature,
484(7394):381–385, 2012.
Joshua I Glaser, Bradley M Zamft, Adam H Marblestone, Jeffrey R
Moffitt, Keith Tyo, Edward S Boyden, George Church, and Konrad P
Kording. Statistical analysis of molecular signal recording. PLOS
Computational Biology, 9(7):e1003145, 2013.
Konrad P Kording. Of toasters and molecular ticker tapes. PLoS
computational biology, 7(12):e1002291, 2011.
Takashi Kawashima, Kazuo Kitamura, Kanzo Suzuki, Mio Nonaka,
Satoshi Kamijo, Sayaka Takemoto-Kimura, Masanobu Kano, Hiroyuki
Okuno, Kenichi Ohki, and Haruhiko Bito. Functional labeling of neurons
and their projections using the synthetic activity-dependent promoter
e-sare. Nature methods, 2013.
J Patrick Card and Lynn W Enquist. Use and visualization of
neuroanatomical viral transneuronal tracers. In Visualization Techniques,
pages 225–268. Springer, 2012.
Evan H Feinberg, Miri K VanHoven, Andres Bendesky, George Wang,
Richard D Fetter, Kang Shen, and Cornelia I Bargmann. Gfp
reconstitution across synaptic partners (grasp) defines cell contacts and
synapses in living nervous systems. Neuron, 57(3):353–363, 2008.
Chaehyun Yook, Shaul Druckmann, and Jinhyun Kim. Mapping
mammalian synaptic connectivity. Cellular and Molecular Life Sciences,
pages 1–11, 2013.
Pavel Osten and Troy W Margrie. Mapping brain circuitry with a light
microscope. Nature methods, 10(6):515–523, 2013.
Kwanghun Chung, Jenelle Wallace, Sung-Yon Kim,
Sandhiya Kalyanasundaram, Aaron S Andalman, Thomas J Davidson,
Julie J Mirzabekov, Kelly A Zalocusky, Joanna Mattis, Aleksandra K
Denisin, et al. Structural and molecular interrogation of intact biological
systems. Nature, 497(7449):332–337, 2013a.
Moritz Helmstaedter. Cellular-resolution connectomics: challenges of
dense neural circuit reconstruction. Nature methods, 10(6):501–507, 2013.
Lukasz J Bugaj, Atri T Choksi, Colin K Mesuda, Ravi S Kane, and
David V Schaffer. Optogenetic protein clustering and signaling activation
in mammalian cells. Nature methods, 2013.
Matthew J Kennedy, Robert M Hughes, Leslie A Peteya, Joel W
Schwartz, Michael D Ehlers, and Chandra L Tucker. Rapid
blue-light-mediated induction of protein interactions in living cells. Nature
methods, 7(12):973–975, 2010.
Dongjin Seo, Jose M Carmena, Jan M Rabaey, Elad Alon, and Michel M
Maharbiz. Neural dust: an ultrasonic, low power solution for chronic
brain-machine interfaces. arXiv preprint arXiv:1307.2196, 2013.
Brian A Wilt, Laurie D Burns, Eric Tatt Wei Ho, Kunal K Ghosh,
Eran A Mukamel, and Mark J Schnitzer. Advances in light microscopy
for neuroscience. Annual review of neuroscience, 32:435, 2009.
Katrin Amunts, Claude Lepage, Louis Borgeat, Hartmut Mohlberg,
Timo Dickscheid, Marc-Étienne Rousseau, Sebastian Bludau, Pierre-Louis
Bazin, Lindsay B Lewis, Ana-Maria Oros-Peusquens, et al. Bigbrain: An
ultrahigh-resolution 3d human brain model. Science, 340(6139):1472–1475,
2013.
Rodrigo Gómez-Martínez, Alberto M Hernández-Pinto, Marta
Duch, Patricia Vázquez, Kirill Zinoviev, J Enrique, Jaume Esteve, Teresa
Suárez, and José A Plaza. Silicon chips detect intracellular pressure
changes in living cells. Nature nanotechnology, 8(7):517–521, 2013.
Michał Horodecki and Jonathan Oppenheim. Fundamental limitations
for quantum and nanoscale thermodynamics. Nature Communications, 4,
2013.
Meng-Tsen Ke, Satoshi Fujimoto, and Takeshi Imai. Seedb: a simple
and morphology-preserving optical clearing agent for neuronal circuit
reconstruction. Nature neuroscience, 2013.
Kwanghun Chung, Jenelle Wallace, Sung-Yon Kim,
Sandhiya Kalyanasundaram, Aaron S Andalman, Thomas J Davidson,
Julie J Mirzabekov, Kelly A Zalocusky, Joanna Mattis, Aleksandra K
Denisin, et al. Structural and molecular interrogation of intact biological
systems. Nature, 497(7449):332–337, 2013b.
Peter A Santi. Light sheet fluorescence microscopy a review. Journal of
Histochemistry & Cytochemistry, 59(2):129–138, 2011.
Peter G Pitrone, Johannes Schindelin, Luke Stuyvenberg, Stephan
Preibisch, Michael Weber, Kevin W Eliceiri, Jan Huisken, and Pavel
Tomancak. Openspim: an open-access light-sheet microscopy platform.
Nature methods, 2013.
Emilio J Gualda, Tiago Vale, Pedro Almada, José A Feijó, Gabriel G
Martins, and Nuno Moreno. Openspinmicroscopy: an open-source
integrated microscopy platform. Nature methods, 2013.
Po-Ru Loh, Michael Baym, and Bonnie Berger. Compressive genomics.
Nature biotechnology, 30(7):627–630, 2012.
Jeffrey Dean and Sanjay Ghemawat. Mapreduce: simplified data
processing on large clusters. Communications of the ACM, 51(1):107–113,
2008.
Michael A Quail, Miriam Smith, Paul Coupland, Thomas D Otto,
Simon R Harris, Thomas R Connor, Anna Bertoni, Harold P Swerdlow,
and Yong Gu. A tale of three next generation sequencing platforms:
comparison of ion torrent, pacific biosciences and illumina miseq
sequencers. BMC genomics, 13(1):341, 2012.
Nicholas J Loman, Raju V Misra, Timothy J Dallman, Chrystala
Constantinidou, Saheer E Gharbia, John Wain, and Mark J Pallen.
Performance comparison of benchtop high-throughput sequencing
platforms. Nature biotechnology, 30(5):434–439, 2012.
Travis C Glenn. Field guide to next-generation dna sequencers.
Molecular Ecology Resources, 11(5):759–769, 2011.
Elizabeth S Sattely, Michael A Fischbach, and Christopher T Walsh.
Total biosynthesis: in vitro reconstitution of polyketide and nonribosomal
peptide pathways. Natural product reports, 25(4):757–793, 2008.
Bradley S Evans, Yunqiu Chen, William W Metcalf, Huimin Zhao,
and Neil L Kelleher. Directed evolution of the nonribosomal peptide
synthetase admk generates new andrimid derivatives in vivo. Chemistry
& biology, 18(5):601–607, 2011.
Bradley W Langhorst, William E Jack, Linda Reha-Krantz, and
Nicole M Nichols. Polbase: a repository of biochemical, genetic and
structural information about dna polymerases. Nucleic acids research, 40
(D1):D381–D387, 2012.
Dong Cai, Lu Ren, Huaizhou Zhao, Chenjia Xu, Lu Zhang, Ying Yu,
Hengzhi Wang, Yucheng Lan, Mary F Roberts, Jeffrey H Chuang, et al.
A molecular-imprint nanosensor for ultrasensitive detection of proteins.
Nature nanotechnology, 5(8):597–601, 2010.
Jonas Korlach, Patrick J Marks, Ronald L Cicero, Jeremy J Gray,
Devon L Murphy, Daniel B Roitman, Thang T Pham, Geoff A Otto,
Mathieu Foquet, and Stephen W Turner. Selective aluminum passivation
for targeted immobilization of single dna polymerase molecules in
zero-mode waveguide nanostructures. Proceedings of the National Academy
of Sciences, 105(4):1176–1181, 2008.
JT Mannion and HG Craighead. Nanofluidic structures for single
biomolecule fluorescent detection. Biopolymers, 85(2):131–143, 2007.
Harold Craighead. Future lab-on-a-chip technologies for interrogating
individual molecules. Nature, 442(7101):387–393, 2006.
John J Kasianowicz, Joseph WF Robertson, Elaine R Chan, Joseph E
Reiner, and Vincent M Stanford. Nanoscopic porous sensors. Annu. Rev.
Anal. Chem., 1:737–766, 2008.
Nan Zhang, Xiaodi Su, Paul Free, Xiaodong Zhou, Koon Gee Neoh,
Jinghua Teng, and Wolfgang Knoll. Plasmonic metal nanostructure array
by glancing angle deposition for biosensing application. Sensors and
Actuators B: Chemical, 2013.
Michael J Heller, Benjamin Sullivan, Dietrich Dehlinger, and Paul
Swanson. Next-generation dna hybridization and self-assembly
nanofabrication devices. In Springer Handbook of Nanotechnology, pages
389–401. Springer, 2010.
Seung Kyu Min, Woo Youn Kim, Yeonchoo Cho, and Kwang S Kim.
Fast dna sequencing with a graphene-based nanochannel device. Nature
nanotechnology, 6(3):162–165, 2011.
Jiayuan Quan, Ishtiaq Saaem, Nicholas Tang, Siying Ma, Nicolas Negre,
Hui Gong, Kevin P White, and Jingdong Tian. Parallel on-chip gene
synthesis and application to optimization of protein expression. Nature
biotechnology, 29(5):449–452, 2011.
K Seidl, S Spieth, S Herwik, J Steigert, R Zengerle, O Paul, and
P Ruther. In-plane silicon probes for simultaneous neural recording and
drug delivery. Journal of Micromechanics and Microengineering, 20(10):
105006, 2010.
Vikram C Sundar, Andrew D Yablon, John L Grazul, Micha Ilan,
Joanna Aizenberg, et al. Fibre-optical features of a glass sponge. Nature,
424(6951):899–900, 2003.
Vikash Gilja, Paul Nuyujukian, Cindy A Chestek, John P Cunningham,
M Yu Byron, Joline M Fan, Mark M Churchland, Matthew T Kaufman,
Jonathan C Kao, Stephen I Ryu, et al. A high-performance neural
prosthesis enabled by control algorithm design. Nature neuroscience, 2012.
Lakshminarayan Srinivasan, Uri T Eden, Sanjoy K Mitter, and
Emery N Brown. General-purpose filter design for neural prosthetic
devices. Journal of Neurophysiology, 98(4):2456–2475, 2007.
Audrey S Royer, Alexander J Doud, Minn L Rose, and Bin He. Eeg
control of a virtual helicopter in 3-dimensional space using intelligent
control strategies. Neural Systems and Rehabilitation Engineering, IEEE
Transactions on, 18(6):581–589, 2010.
Alexander J Doud, John P Lucas, Marc T Pisansky, and Bin He.
Continuous three-dimensional control of a virtual helicopter using a motor
imagery based brain-computer interface. PloS one, 6(10):e26322, 2011.
Todd F Roberts, Sharon MH Gobes, Malavika Murugan, Bence P
Ölveczky, and Richard Mooney. Motor circuits are required to encode a
sensory model for imitative learning. Nature Neuroscience, 2012.
Goded Shahaf and Shimon Marom. Learning in networks of cortical
neurons. The Journal of Neuroscience, 21(22):8782–8788, 2001.
Sophie Pautot, Claire Wyart, and Ehud Y Isacoff. Colloid-guided
assembly of oriented 3d neuronal networks. Nature methods, 5(8):735–740,
2008.
Bruce C Wheeler and Gregory J Brewer. Designing neural networks in
culture. Proceedings of the IEEE, 98(3):398–406, 2010.
Saul A Villeda, Jian Luo, Kira I Mosher, Bende Zou, Markus Britschgi,
Gregor Bieri, Trisha M Stan, Nina Fainberg, Zhaoqing Ding, Alexander
Eggel, et al. The ageing systemic milieu negatively regulates neurogenesis
and cognitive function. Nature, 477(7362):90–94, 2011.
Hartmuth C Kolb, MG Finn, and K Barry Sharpless. Click chemistry:
diverse chemical function from a few good reactions. Angewandte Chemie
International Edition, 40(11):2004–2021, 2001.
R Feil, J Brocard,
B Mascrez, M LeMeur, D Metzger, and P Chambon. Ligand-activated
site-specific recombination in mice. Proceedings of the National Academy
of Sciences, 93(20):10887–10890, 1996.
Larry J Kricka. Microchips, microarrays, biochips and nanochips:
personal laboratories for the 21st century. Clinica Chimica Acta, 307(1):
219–223, 2001.
Guijian Guan, Bianhua Liu, Zhenyang Wang, and Zhongping Zhang.
Imprinting of molecular recognition sites on nanostructures and its
applications in chemosensors. Sensors, 8(12):8291–8320, 2008.
Robi D Mitra, Jay Shendure, Jerzy Olejnik, George M Church, et al.
Fluorescent in situ sequencing on polymerase colonies. Analytical
biochemistry, 320(1):55–65, 2003.
Mikhail G Shapiro, Gil G Westmeyer, Philip A Romero, Jerzy O
Szablowski, Benedict Küster, Ameer Shah, Christopher R Otey, Robert
Langer, Frances H Arnold, and Alan Jasanoff. Directed evolution of a
magnetic resonance imaging contrast agent for noninvasive imaging of
dopamine. Nature biotechnology, 28(3):264–270, 2010.
Eric M Brustad, Victor S Lelyveld, Christopher D Snow, Nathan Crook,
Sang Taek Jung, Francisco M Martinez, Timothy J Scholl, Alan Jasanoff,
and Frances H Arnold. Structure-guided directed evolution of highly
selective p450-based magnetic resonance imaging sensors for dopamine and
serotonin. Journal of Molecular Biology, 2012.
Binyamin Hochner. Octopus nervous system. Encyclopedia of
Neuroscience, 2004.
Loretta Guidi, Michael Eitel, Erica Cesarini, Bernd Schierwater, and
Maria Balsamo. Ultrastructural analyses support different morphological
lineages in the phylum placozoa grell, 1971. Journal of Morphology, 272
(3):371–378, 2011.
Jeffrey H Ringrose, Henk WP van den Toorn, Michael Eitel, Harm Post,
Pieter Neerincx, Bernd Schierwater, AF Maarten Altelaar, and Albert JR
Heck. Deep proteome profiling of trichoplax adhaerens reveals remarkable
features at the origin of metazoan multicellularity. Nature communications,
4:1408, 2013.
Wolfgang Jakob, Sven Sagasser, Stephen Dellaporta, Peter Holland,
Kerstin Kuhn, and Bernd Schierwater. The trox-2 hox/parahox gene of
trichoplax (placozoa) marks an epithelial boundary. Development genes
and evolution, 214(4):170–175, 2004.
Michael Eitel, Loretta Guidi, Heike Hadrys, Maria Balsamo, and Bernd
Schierwater. New insights into placozoan sexual reproduction and
development. PLoS One, 6(5):e19639, 2011.
Olivia Mendivil Ramos, Daniel Barker, and David EK Ferrier. Ghost
loci imply hox and parahox existence in the last common ancestor of
animals. Current Biology, 2012.
Mansi Srivastava, Emina Begovic, Jarrod Chapman, Nicholas H Putnam,
Uffe Hellsten, Takeshi Kawashima, Alan Kuo, Therese Mitros, Asaf
Salamov, Meredith L Carpenter, et al. The trichoplax genome and the
nature of placozoans. Nature, 454(7207):955–960, 2008.
Franz Eilhard Schulze. Über Trichoplax adhaerens. Verlag d. Königl.
Akad. d. Wiss., 1892.
KG Grell. Trichoplax adhaerens fe schulze und die entstehung der
metazoen. Naturwiss. Rundschau, 24(4):160–161, 1971.
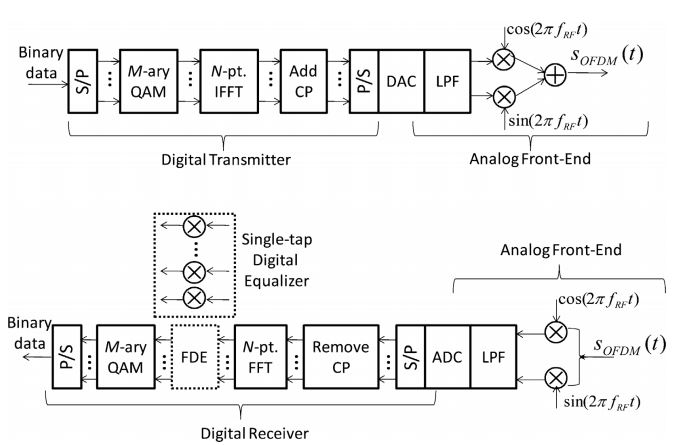
Dayou Qian, Ming-Fang Huang, Ezra Ip, Yue-Kai Huang, Yin Shao,
Junqiang Hu, and Ting Wang. High capacity/spectral efficiency 101.7-tb/s
wdm transmission using pdm-128qam-ofdm over 165-km ssmf within c-and
l-bands. Journal of Lightwave Technology, 30(10):1540–1548, 2012.
Neda Cvijetic. Ofdm for next-generation optical access networks.
Lightwave Technology, Journal of, 30(4):384–398, 2012.
Madeleine Glick. Optical interconnects in next generation data centers:
an end to end view. In Optical Interconnects for Future Data Center
Networks, pages 31–46. Springer, 2013.
Elizabeth Jurrus, Melissa Hardy, Tolga Tasdizen, P Thomas Fletcher,
Pavel Koshevoy, Chi-Bin Chien, Winfried Denk, and Ross Whitaker. Axon
tracking in serial block-face scanning electron microscopy. Medical image
analysis, 13(1):180–188, 2009.
H Sebastian Seung. Neuroscience: towards functional connectomics.
Nature, 471(7337):170–172, 2011.
Aaron McKenna, Matthew Hanna, Eric Banks, Andrey Sivachenko,
Kristian Cibulskis, Andrew Kernytsky, Kiran Garimella, David Altshuler,
Stacey Gabriel, Mark Daly, et al. The genome analysis toolkit: a
mapreduce framework for analyzing next-generation dna sequencing data.
Genome research, 20(9):1297–1303, 2010.
K Ingemar Jönsson, Elke Rabbow, Ralph O Schill,
Mats Harms-Ringdahl, and Petra Rettberg. Tardigrades survive exposure
to space in low earth orbit. Current biology, 18(17):R729–R731, 2008.
Gunther S Stent, William B Kristan Jr, W Otto Friesen, Carol A Ort,
Margaret Poon, and Ronald L Calabrese. Neuronal generation of the leech
swimming movement. Science, 200(4348):1348–1357, 1978.
’Smart genes’ prove elusive, 1
., 2
‘the brain is hot’, 3
20 Years of Computational Neuroscience, 4, 5
2015 preview: First human trials to cure blindness, 6
457.02 - The impact of pain on motivation, 7
457.05 - The anterior cingulate cortex as a substrate for chronic pain-induced depression: molecular, lesional and optogenetic evidences., 8
457.06 - Nucleus Accumbens subregions dissociate encoding of values for reward and pain, 9
457.07 - Intracellular pathways in the Nucleus Accumbens modulate the antiallodynic actions of antidepressants., 10
5 Ways to Adjust to Graduate School, 11
549.02 - Optogenetic control of pain and motor circuitry, 12
549.03 - Optical control of stem cell derived motor neurons restores function to paralysed muscles, 13
549.04 - New paradigms in wireless light delivery, 14
549.05 - An Optogenetic Demonstration of Motor Primitives in the Mouse Spinal Cord, 15
549.06 - Optogenetic control of aversive sensory circuitry, 16
549.07 - Optogenetic dissection of visceral pain, 17
739.06 - Chronic pain causes dysfunction in reward circuitry, 18
739.07 - Distinct subpopulations of dynorphin neurons drive aversion and reward, 19
9.14 - Brain Structure and Its Origins, 20
9.29J, 21
‘Silent’ mitral cells dominate odor responses in the olfactory bulb of awake mice, 22
A comprehensive thalamocortical projection map at the mesoscopic level, 23, 24
A mesoscale connectome of the mouse brain, 25
A million spiking-neuron integrated circuit with a scalable communication network and interface, 26
A synaptic organizing principle for cortical neuronal groups, 27
A Whole-Brain Atlas of Inputs to Serotonergic Neurons of the Dorsal and Median Raphe Nuclei, 28
A Whole-Brain Atlas of Inputs to Serotonergic Neurons of the Dorsal and Median Raphe Nuclei , 29
Alone in the Void , 30
Anatomy of Scientific Evolution, 31
BRAIN: Launching America^^e2^^80^^99s Next Moonshot, 32
Can Humans Really Discriminate 1 Trillion Odors? , 33
Career on the Move: Geography, Stratification, and Scientific Impact, 34
Centauri Dreams: Imagining and Planning Interstellar Exploration, 35
Cerebellum involvement in cortical sensorimotor circuits for the control of voluntary movements, 36
Computer-Generated Holographic Beams for the Investigation of the Molecular and Circuit Function, 37
Constraining the connectivity of neuronal networks cultured on microelectrode arrays with microfluidic techniques: A step towards neuron-based functional chips, 38
Cosmos: A Personal Voyage , 39
Creating effective slides: Design, Construction, and Use in Science (youtube) , 40
current opinion in neurobiology: focus on circuits, 41
Deep Space Propulsion: A Roadmap to Interstellar Flight, 42
Development, learning and memory in large random networks of cortical neurons: lessons beyond anatomy, 43
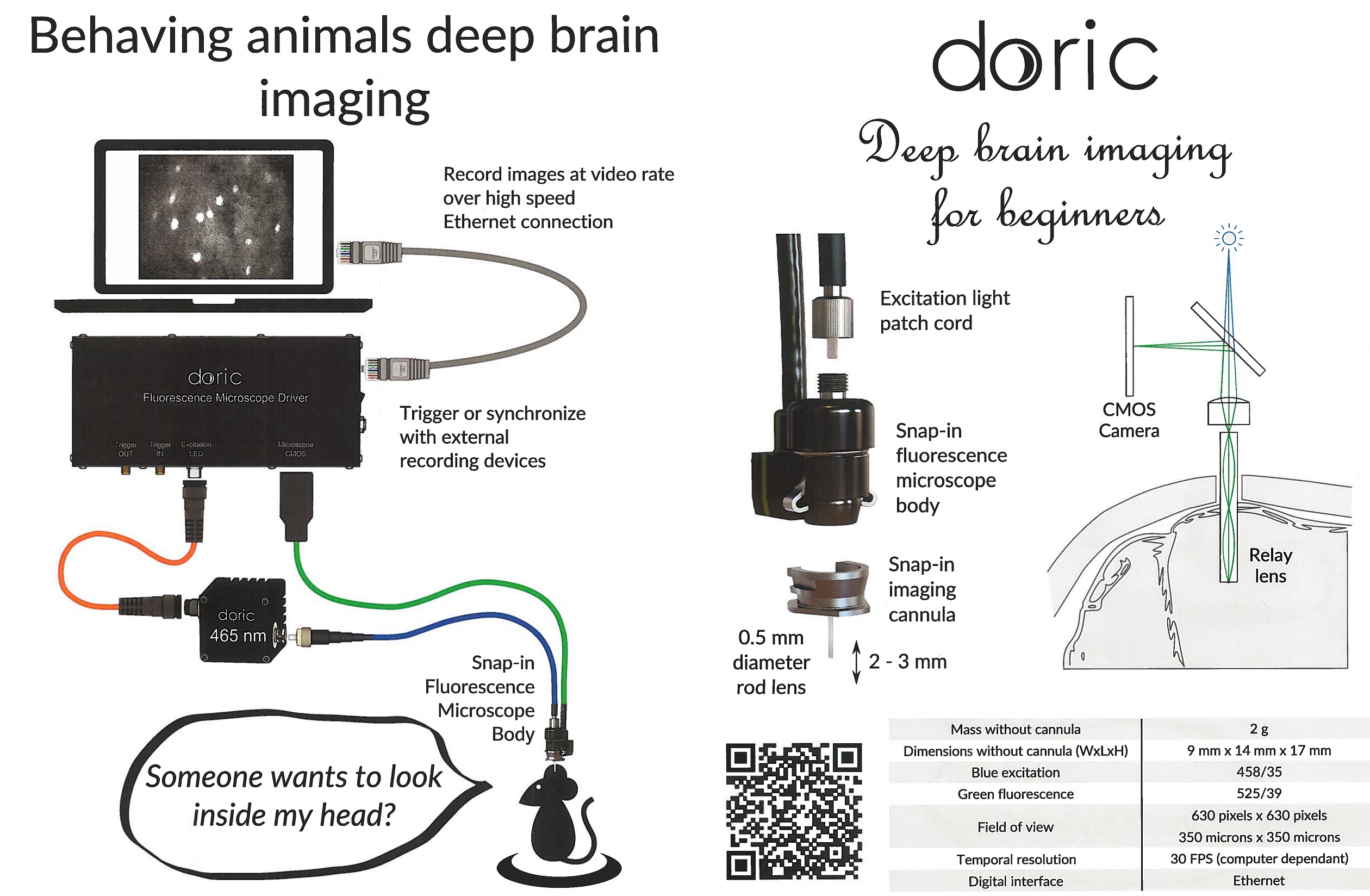
Doric Miniaturized Fluorescence Microscopy, 44
Driving forces of researchers mobility, 45
Duarte Portfolio , 46
Effective Presentations in Engineering and Science, 47
Entering Space, 48
Functional patterned multiphoton excitation deep inside scattering tissue, 49
Fundamentals of Astrodynamics , 50
Future impact: Predicting scientific success, 51
Future Science, 52
Genome-scale functional characterization of Drosophila developmental enhancers in vivo , 53
Giving science talks , 54
http://noteandpoint.com/ , 55
Humans Can Discriminate More than 1 Trillion Olfactory Stimuli , 56
Identifying Functional Connections of the Inner Photoreceptors in Drosophila using Tango-Trace, 57
Imaging Activity in Neurons and Glia with a Polr2a-Based and Cre-Dependent GCaMP5G-IRES-tdTomato Reporter Mouse , 58
Improve your PowerPoint (youtube), 59
Interstellar travel (wikipedia), 60
Interstellar Travel andMulti-Generational Space Ships: Apogee Books Space Series 34, 61
Islands in the Sky: Bold New Ideas for Colonizing Space, 62
Learning in Networks of Cortical Neurons, 63
Light Field Microscopy for 3D functional neuroimaging, 64
Limits on fundamental limits to computation, 65
Long-Term Temporal Imprecision of Information Coding in the Anterior Cingulate Cortex of Mice with Peripheral Inflammation or Nerve Injury, 66
microcircuits and their computational roles, 67
Mining The Sky: Untold Riches From The Asteroids, Comets, And Planets, 68
Multiscale Optical Ca2+ Imaging of Tonal Organization in Mouse Auditory Cortex , 69
Network Structure within the Cerebellar Input Layer Enables Lossless Sparse Encoding , 70
Neural Networks of the Mouse Neocortex, 71
Neurons Are Recruited to a Memory Trace Based on Relative Neuronal Excitability Immediately before Training, 72
Neuroscience Technologies, 73
NIH awards initial $46 million for BRAIN Initiative research, 74
On the Predictability of Future Impact in Science, 75
Organization of Monosynaptic Inputs to the Serotonin and Dopamine Neuromodulatory Systems , 76
Pimp your PowerPoint , 77
Prediction of highly cited papers, 78
Presynaptic Partners of Dorsal Raphe Serotonergic and GABAergic Neurons, 79
Proposal For Neuromorphic Hardware Using Spin Devices, 80
Quantifying long-term scientific impact, 81
resonate - Present Visual Stories that Transform Audiences, 82
Robust multicellular computing using genetically encoded NOR gates and chemical ‘wires’, 83
Scientific Presentations The Assertion-Evidence Approach , 84
Sensory-Related Neural Activity Regulates the Structure of Vascular Networks in the Cerebral Cortex, 85
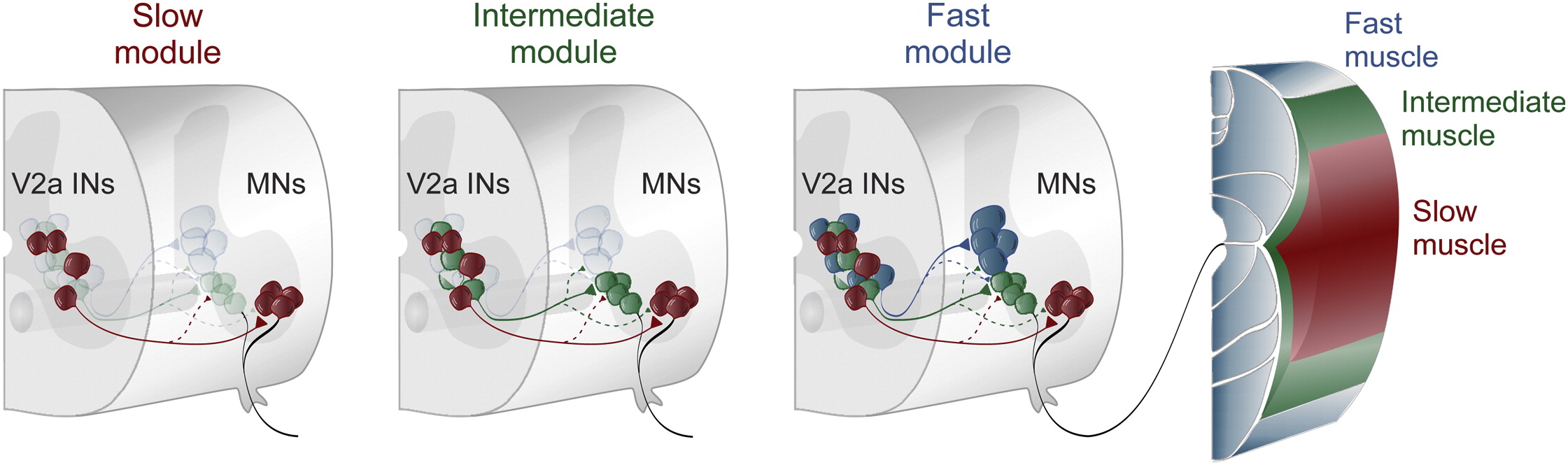
Separate Microcircuit Modules of Distinct V2a Interneurons and Motoneurons Control the Speed of Locomotion, 86
Simultaneous cellular-resolution optical perturbation and imaging of place cell firing fields, 87
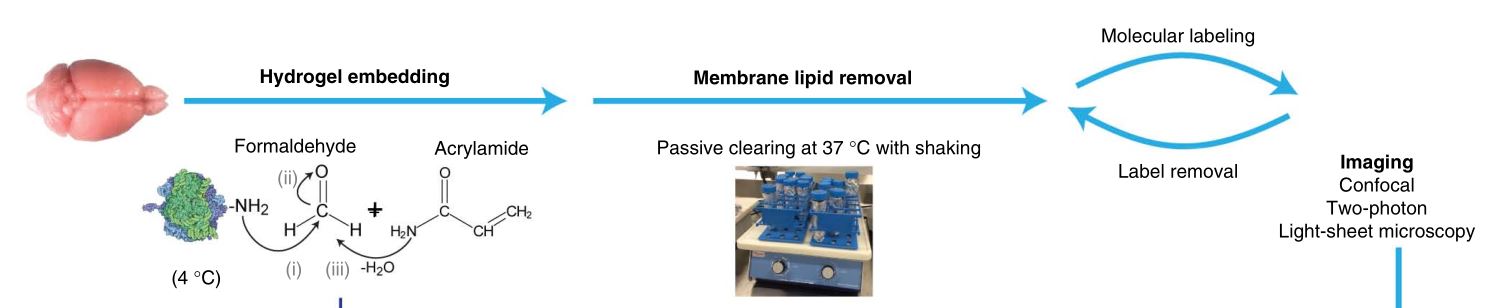
Single-Cell Phenotyping within Transparent Intact Tissue through Whole-Body Clearing, 88
slide:ology: The Art and Science of Creating Great Presentations, 89
Spacefaring: The Human Dimension, 90
Sparseness and Expansion in Sensory Representations, 91
Susan McConnell (Stanford): Designing effective scientific presentations (youtube) , 92
Synaptic plasticity in micropatterned neuronal networks, 93
Targeting neurons and photons for optogenetics , 94
The Case for Mars: The Plan to Settle the Red Planet and Why We Must, 95
The Craft of Scientific Presentations: Critical Steps to Succeed and Critical Errors to Avoid , 96
the Martian , 97
The Presentation Secrets of Steve Jobs, 98
The Starflight Handbook: A Pioneer’s Guide to Interstellar Travel, 99
Thy1-GCaMP6 Transgenic Mice for Neuronal Population Imaging In Vivo, 100
Time to Plan for a Mission to Alpha Centauri , 101
Top Pharma Projects to Watch, 102
Two-photon excitation in scattering media by spatiotemporally shaped beams and their application in optogenetic stimulation, 103
Use of differentiated pluripotent stem cells as replacement therapy for treating disease, 104
Visualizing mammalian brain area interactions by dual-axis two-photon calcium imaging, 105
We Have Met the Enemy and He Is PowerPoint , 106
Wide field-of-view, twin-region two-photon imaging across extended cortical networks, 107
A brain in a supercomputer, 108
A brief account of nanoparticle contrast agents for photoacoustic imaging, 109
A canonical microcircuit for neocortex., 110
A Causative Link Between Inner Ear Defects and Long-Term Striatal Dysfunction, 111
A comprehensive multiscale framework for simulating optogenetics in the heart, 112
A computational perspective on the neural basis of multisensory spatial representations, 113
A database of Caenorhabditis elegans behavioral phenotypes, 114
A Direct Brain-to-Brain Interface in Humans, 115
A Family of Algorithms for Computing Consensus about Node State from Network Data, 116
A faster, brighter picture of brain cells in action, 117
A gene complex controlling segmentation in Drosophila, 118
A graduate school survival guide: “So long, and thanks for the Ph.D
”, 119
A Guide to In vivo Single-unit Recording from Optogenetically Identified Cortical Inhibitory Interneurons, 120
A learning algorithm for Boltzmann machines., 121
A lingering smell?, 122
A marker induction mechanism for the establishment of ordered neural mappings: its application to the retinotectal problem, 123
A Miniature Head-Mounted Two-Photon Microscope: High-Resolution Brain Imaging in Freely Moving Animals, 124
A model for the formation of orientation columns, 125
A neuronal network for computing population vectors in the leech, 126
A Physicist’s Renewed Look at Biology, 127
a previous post, 128
A quantitative description of membrane current and its application to conduction and excitation in nerve, 129
A Robot Really Committed A Crime: Now What?, 130
A Rulebook for Arguments, 131
A self-organizing neural network that discovers surfaces in random-dot stereograms, 132
A simple coding procedure enhances a neuron’s information capacity., 133
A Skeptic’s Guide to the Mind, 134
A spinal analog of memory reconsolidation enables reversal of hyperalgesia, 135
A Standard for Neuroscience Data, 136
A Wireless Multi-Channel Recording System for Freely Behaving Mice and Rats, 137
AA35 - Two-photon imaging of light-induced nociceptive processing In vivo, 138
Abstract - FENS Forum 2014, 139
Accelerando, 140
Accelerated chemistry in the reaction between the hydroxyl radical and methanol at interstellar temperatures facilitated by tunnelling, 141
Activation of the renin-angiotensin system, specifically in the subfornical organ is sufficient to induce fluid intake., 142
Adaptive informatics for multifactorial and high-content biological data, 143
Addressing the Nation^^e2^^80^^99s Changing Needs for Biomedical and Behavioral Scientists, 144
Advanced CLARITY for rapid and high-resolution imaging of intact tissues, 145
advances in artificial intelligence, 146
advances in artificial intelligence, part 2, 147
Advances in the pharmacological treatment of Parkinson’s disease: targeting neurotransmitter systems, 148
Advances in using MRI probes and sensors for in vivo cell tracking as applied to regenerative medicine, 149
Advancing Genetic Treatments Against Blindness, 150
Age and scientific productivity. Differences between fields of learning, 151
Age and the Nobel prize revisited, 152
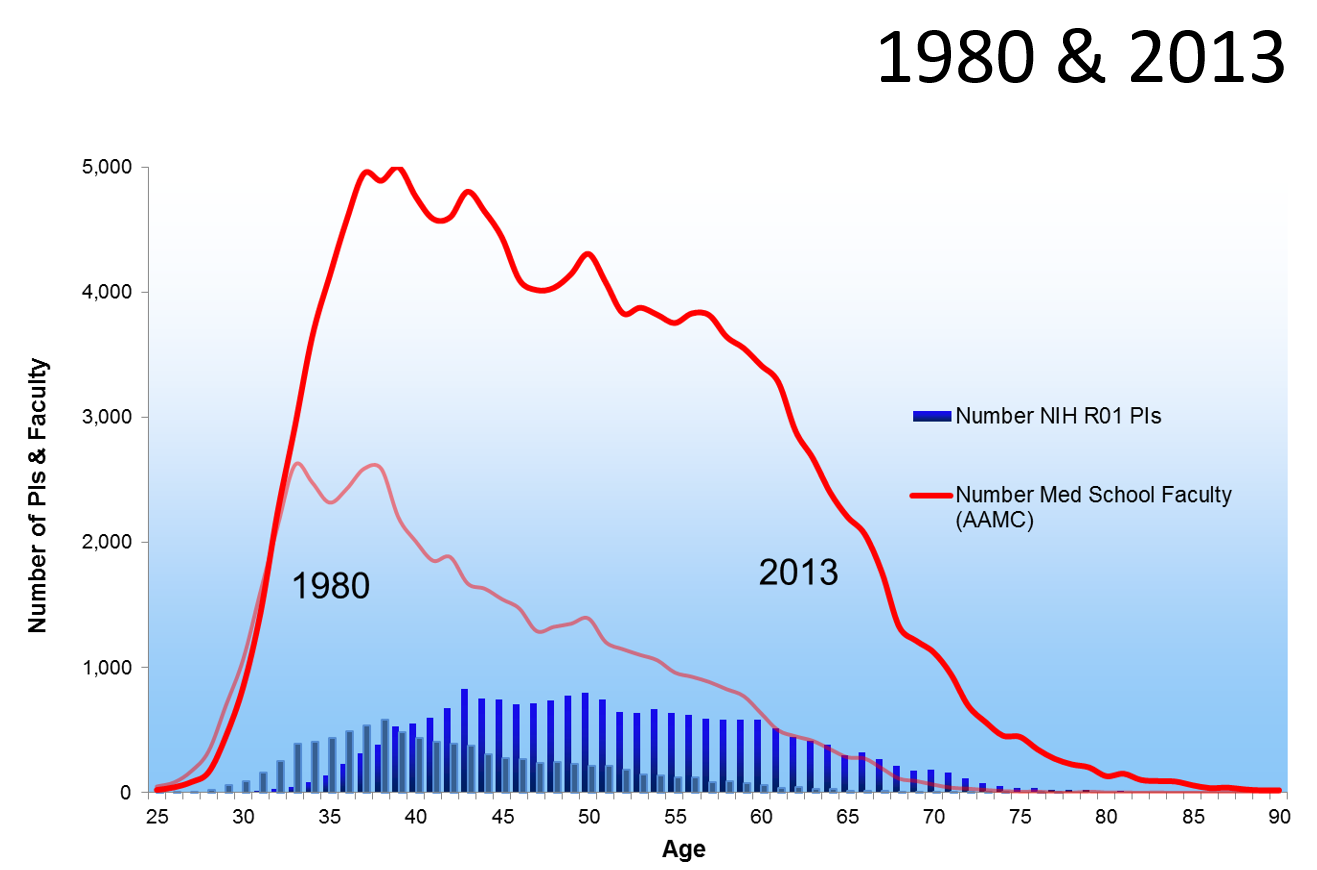
Age Distribution of NIH Principal Investigators and Medical School Faculty, 153
age-related macular degeneration, 154
Ahrens, 2013, 155
Air France Flight 447, 156
Akerboom, 2013, 157
Alan Jasanoff, 158, 159
Albert Lee’s work looking at silent place cells, 160
All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins, 161
All-Optical Interrogation of Neural Circuits, 162
Allelic heterogeneity in NCF2 associated with systemic lupus erythematosus (SLE) susceptibility across four ethnic populations, 163
Allen Brain Atlas, 164
Allen Cell Types Database, 165
Allen Institute, 166, 167
Allen Institute Brain Explorer, 168
along with others, 169
Alzheimer’s trials, 170
amazing work with birds, 171
Ambition the film , 172
Amy Wagers, 173
Amygdala interneuron subtypes control fear learning through disinhibition, 174
An active membrane model of the cerebellar Purkinje cell. I. Simulation of current clamps in slice, 175
An analogue approach to the travelling salesman problem using an elastic net method., 176
An electrical investigation of effects of repetitive stimulation on mammalian neuromuscular junction, 177
An exploratory test for an excess of significant findings, 178
An Interactive Resource to Identify Cancer Genetic and Lineage Dependencies Targeted by Small Molecules, 179
An ultra-lightweight design for imperceptible plastic electronics, 180
Analogous responses in the nucleus accumbens and cingulate cortex to pain onset (aversion) and offset (relief) in rats and humans, 181
Analysis of Transduction Efficiency, Tropism and Axonal Transport of AAV Serotypes 1, 2, 5, 6, 8 and 9 in the Mouse Brain, 182
Ancestral dichlorodiphenyltrichloroethane (DDT) exposure promotes epigenetic transgenerational inheritance of obesity, 183
Anderson, 184
Andre Esteva, 185
Angiotensin, Thirst, and Sodium Appetite, 186
Angiotensinergic and cholinergic receptors of the subfornical organ mediate sodium intake induced by GABAergic activation of the lateral parabrachial nucleus., 187
Annual Review of Neuroscience, 188
Annual Reviews, 189
Anteroventral Wall Of The Third Ventricle And Dorsal Lamina Terminalis: Headquarters For Control Of Body Fluid Homeostasis?, 190
Aping Mankind, 191
Application of Tissue Clearing and Light Sheet Fluorescence Microscopy to Assess Optic Nerve Regeneration in Unsectioned Tissues, 192
Are two heads better than one? The psychology of Pacific Rim, 193
array tomography, 194
article 1, 195
article 2, 196
article 3, 197
artificial intelligence and the law, 198
Artificial intelligence: Learning to see and act, 199
Arvix, 200, 201
arXiv has author identifiers, 202
Assignment of Model Amygdala Neurons to the Fear Memory Trace Depends on Competitive Synaptic Interactions, 203
At what age do biomedical scientists do their best work?, 204
Auditory spatial receptive fields created by multiplication, 205
Autism: A long genetic explanation, 206
Autodesk Inventor, 207
Autodesk’s Inventor, 208
Automated whole-cell patch clamp electrophysiology of neurons in vivo, 209
Autonomous molecular cascades for evaluation of cell surfaces, 210
Autonomous screening of C. elegans identifies genes implicated in synaptogenesis, 211
Autopatcher, 212
awesome work, 213
Axonal delay lines for time measurement in the owl’s brainstem, 214
Bacteria activate sensory neurons that modulate pain and inflammation, 215
Bacteria get on your nerves, 216
bahanonu.com associated blog post, 217
Balanced cortical microcircuitry for maintaining information in working memory, 218
Barabasi, 219
Barabasi Lab, 220
Bargmann, 221
Barney Cruz, 222
Barthas 2015, 223
Basic and Applied Social Psychology Editorial on the null hypothesis significance testing procedure, 224
BB9 - Epidural optic fiber implant for spinal optogenetics, 225
Becerra, 2013, 226
Behavioural battery testing: Evaluation and behavioural outcomes in 8 inbred mouse strains, 227
Ben Barres, 228
Benjamin Grewe, 229
Between Postdoc and Job, a Whole Lot of Questions, 230
BGI Cognitive Genomics, 231
Bidirectional switch of the valence associated with a hippocampal contextual memory engram, 232
Big data from small data: data-sharing in the ‘long tail’ of neuroscience, 233
Big data, but are we ready?, 234
Big Science vs. Little Science: How Scientific Impact Scales with Funding, 235
Binless strategies for estimation of information from neural data, 236
Biophysical model of a Hebbian synapse, 237
Bit-by-bit autophagic removal of parkin-labelled mitochondria, 238
BlackRock, 239
Blocking receptor in brain^^e2^^80^^99s immune cells counters Alzheimer^^e2^^80^^99s in mice, study finds, 240
Blue Brain project, 241
blue brain project, 242
Bongwoori, 243
Boyden, 244, 245
Boyden Lab, 246
BRAIN 2025: A Scientific Vision, 247
Brain Activity in Valuation Regions while Thinking about the Future Predicts Individual Discount Rates, 248
brain bank, 249
Brain Computation as Hierarchical Abstraction, 250
BRAIN initiative, 251, 252, 253
brain initiative notes, 254, 255
brain initiative notes rss feed, 256
BRAIN short story, 257
Brain Windows, 258
brain-computer interfaces, 259
Brain-machine interfaces, 260
Brain/MINDS: brain-mapping project in Japan, 261
BrainAligner, 262
Brainbow, 263
BrainFormat, 264, 265
BrainGate, 266
Branson Lab, 267, 268
Breakdown of long-range temporal dependence in default mode and attention networks during deep sleep, 269
Breakthrough Therapies, 270
Bridging the transgenerational gap with epigenetic memory, 271
Briggman, 272
Brute force searching, the typical set and Guesswork, 273
Buck Institute, 274, 275, 276
CA3 spine thorns, 277
calcineurin, 278
calcium imaging cell detection techniques, 279
Calcium sensor leaves permanent mark in activated neurons, 280
Callaway, 281
Calling the next generation of affinity reagents, 282
CaMPARI, 283
Campari, 284
Can a biologist fix a radio?^^e2^^80^^94Or, what I learned while studying apoptosis, 285
Can Machines Think?, 286
Can matter cycle through shapes eternally?, 287
Cannabinoids in Pain Management: An Update, 288
Cardiac function and electrical remodeling of the calcineurin-overexpressed transgenic mouse, 289
Casting light on pain, 290
Causes and Consequences of Hyperexcitation in Central Clock Neurons, 291
Cell, 292, 293
cell assemblies as proposed by Buzaki, 294
Cell mechanism overload, 295
Cellular and Synaptic Architecture of Multisensory Integration in the Mouse Neocortex, 296
Cellular organization of cortical barrel columns is whisker-specific, 297
Cellular Resolution Functional Imaging in Behaving Rats Using Voluntary Head Restraint, 298
Cellular resolution optical access to brain regions in fissures: Imaging medial prefrontal cortex and grid cells in entorhinal cortex, 299
Cerebral cartography: a vision of its future, 300
Cerebral organoids model human brain development and microcephaly, 301
CERN uses magnetic tapes for storage still, 302
Chance and consensus in peer review, 303
channelpedia, 304
channelrhodopsin, 305
Channelrhodopsin as a tool to investigate synaptic transmission and plasticity., 306
channels, 307
Characteristic Effects of Stochastic Oscillatory Forcing on Neural Firing: Analytical Theory and Comparison to Paddlefish Electroreceptor Data, 308
check out his lab’s website, 309
Chemical Reviews, 310, 311
Chemogenetic Synaptic Silencing of Neural Circuits Localizes a Hypothalamus-¿Midbrain Pathway for Feeding Behavior, 312
Cheng-Hsun Wu, 313
Chinese scientists genetically modify human embryos, 314
chlorsulfuron, 315
Choosing a Lab, 316
Choosing a Thesis Lab: Things to consider before and during rotations, 317
Choosing The Right Research Adviser, 318
chromatophore, 319
Chronic Cellular Imaging of Entire Cortical Columns in Awake Mice Using Microprisms, 320
chronic, wireless recordings of large-scale brain activity in freely moving rhesus monkeys, 321
Chronos and Chrimson, 322
Chronos and Crimson, 323
Church, 324
Church Lab, 325
churchland lab, 326
Citation opportunity cost of the high impact factor obsession, 327
CLARITY, 328
ClearT, 329
CLEO, 330
click chemistry, 331
CNC Annual Symposium, 332
Cocaine-induced structural plasticity in frontal cortex correlates with conditioned place preference, 333
Cognitive neuroscience: Time, space and memory, 334
Cohen, 335
Collaborative Research in Computational Neuroscience, 336
Combinatorial Mutagenesis of the Voltage-Sensing Domain Enables the Optical Resolution of Action Potentials Firing at 60 Hz by a Genetically Encoded Fluorescent Sensor of Membrane Potential, 337
COMBREX, 338
Comparison of Linux Development Boards, 339
Competition and Careers in Biosciences, 340
CompTop, 341
Computational analysis of the role of the hippocampus in memory, 342
Computational design of ligand-binding proteins with high affinity and selectivity, 343
Computational vision and regularization theory., 344
Computer-Generated Holographic Beams for the Investigation of the Molecular and Circuit Function, 345
Computers Faster Only for 75 More Years, 346
Computing with 10,000-Bit Words*, 347
Conceptual Models and Analytical Tools: The Biology of Physicist Max Delbr^^c3^^bcck, 348
Concurrent Imaging of Synaptic Vesicle Recycling and Calcium Dynamics, 349
Connectomic reconstruction of the inner plexiform layer in the mouse retina, 350
Consciousness: Confessions of a Romantic Reductionist, 351
Consciousness: here, there and everywhere?, 352
Considerations for developing a standard for storing electrophysiology data in HDF5, 353
Constitutive ^^ce^^bc-Opioid Receptor Activity Leads to Long-Term Endogenous Analgesia and Dependence, 354
Conte Center Neuroscience Symposium, 355
context switches, 356
continue to falter, 357
Control VR - The Future of VR and Animation, 358
Conversion of Channelrhodopsin into a Light-Gated Chloride Channel, 359
Cooperative Computation of Stereo Disparity, 360
Correlated neuronal discharge rate and its implications for psychophysical performance, 361
Correlation between neural spike trains increases with firing rate, 362
Cortical connections and parallel processing: Structure and function, in Vision, in Brain and cooperative computation, 363
Cortical development and remapping through spike timing-dependent plasticity, 364
cortical neuron cultures, 365
Cortical rewiring and information storage, 366
Corticostriatal functional connectivity predicts transition to chronic back pain, 367
Cost-Based Metrics for Spike Trains, 368
Could information theory provide an ecological theory of sensory processing?, 369
Coupling Mechanism and Significance of the BOLD Signal: A Status Report, 370
Cox, 371
Creating a False Memory in the Hippocampus, 372
CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes, 373
cs379c, 374, 375, 376, 377, 378, 379
CSHL, 380
CT scans, 381
CUDA, 382
CUDA tutorials, 383
cultured neuron interface, 384
Current Opinion in Neurobiology: Theoretical and computational neuroscience, 385
cyclic voltametry, 386
cycorp, 387
da Vinci surgical system, 388
Daniel Eth, 389
Data Science at NIH, 390
Data Sharing for Computational Neuroscience, 391
Data URL, 392, 393
David Attwell, 394
David Cox, 395, 396
David Hilbert, 397
DBpedia, 398
DD3 - Optogenetic control of dopamine release in rodents and novel opto-dopamine probes for In vivo experiments, 399
decision by consensus, 400
Decisions, decisions, 401, 402
Decoding Information in Cell Shape, 403
Decoding Neural Circuits that Control Compulsive Sucrose Seeking, 404
Decreased motivation during chronic pain requires long-term depression in the nucleus accumbens, 405
DeepMind, 406
FDOT14, 407
Deisseroth lab, 408
Demystifying multithreading and multi-core, 409
Dendrimeric calcium-responsive MRI contrast agents with slow in vivo diffusion, 410
Dendritic computation, 411
dendritic computation, 412
dendritic spines, 413
dephosphorylation, 414
Depression of transmitter release at the neuromuscular junction of the frog, 415
Design Constraints for Mobile, High-Speed Fluorescence Brain Imaging in Awake Animals, 416
designer babies, 417
Desipramine, 418
Detecting cells using non-negative matrix factorization on calcium imaging data, 419
Detrimental effects of reward: Reality or myth?, 420
Development and Applications of CRISPR-Cas9 for Genome Engineering, 421
Devon Chandler-Brown, 422
Dias, 2014, 423
dielectric resonator, 424
Diesseroth, 425
Diesseroth Lab, 426
Differences in Review Quality and Recommendations for Publication Between Peer Reviewers Suggested by Authors or by Editors, 427
Differential signaling via the same axon of neocortical pyramidal neurons, 428
Digimouse: 3D Mouse Atlas, 429, 430
digital object identifier system, 431
Direct visuomotor transformations for reaching, 432
Discovery of Brainwide Neural-Behavioral Maps via Multiscale Unsupervised Structure Learning, 433, 434, 435
Distinct Basal Ganglia Circuits Controlling Behaviors Guided by Flexible and Stable Values, 436
Distinct Representations of Cognitive and Motivational Signals in Midbrain Dopamine Neurons, 437
Distinct Subpopulations of Nucleus Accumbens Dynorphin Neurons Drive Aversion and Reward, 438
DIWG’s executive summary, 439
DNA methylation, 440
DNA methylation regulates associative reward learning, 441
DNA polymerase, 442
Do Animal Models Tell Us about Human Pain?, 443
Do rats have a prefrontal cortex?, 444
Does Google Scholar contain all highly cited documents (1950-2013)?, 445
DOMDocument, 446
DOMXPath, 447
Don^^e2^^80^^99t edit the human germ line, 448
Donoghue, 449
Dopamine Modulates Risk-Taking as a Function of Baseline Sensation-Seeking Trait, 450
dopamine receptors, 451
Dopaminergic Control of Long-Term Depression/Long-Term Potentiation Threshold in Prefrontal Cortex, 452
Dorsal Raphe Neurons Signal Reward through 5-HT and Glutamate, 453
Double Nicking by RNA-Guided CRISPR Cas9 for Enhanced Genome Editing Specificity, 454, 455
Douglas Fields, 456
DP03 - Measurement of phasic dopamine signals in the rat nucleus accumbens core and shell in response to noxious stimuli, 457
Dr. Salzmann’s work at Max Planck Institute for Biological Cybernetics, 458
DREADDs, 459
Drinking behavior following electrical stimulation of the subfornical organ in the rat., 460
Drosophila, 461
Drug development: Raise standards for preclinical cancer research, 462
dual photo-stimulation and imaging, 463, 464, 465, 466
dual photo-stimulation and imaging, cont’d, 467
dual photo-stimulation and imaging, cont^^e2^^80^^99d, 468
Dynamically Reshaping Signaling Networks to Program Cell Fate via Genetic Controllers, 469
Dysfunction of Cortical Dendritic Integration in Neuropathic Pain Reversed by Serotoninergic Neuromodulation, 470
echo state networks, 471
Ecological expected utility and the mythical neural code., 472
Ecosystem consequences of bird declines, 473
Editorial peer review for improving the quality of reports of biomedical studies, 474
Effect on the Quality of Peer Review of Blinding Reviewers and Asking Them to Sign Their Reports, 475
Effect size, 476
Effect size, confidence interval and statistical significance: a practical guide for biologists, 477
Effects of Editorial Peer Review, 478
Effects of training on quality of peer review: randomised controlled trial, 479
electrically tunable lenses, 480
electroencephalography, 481
electron microscopy, 482
Electrophysiology Task Force, 483
Elizabeth M. C. Hillman, 484
Elon Musk - Work ethics, Principles, Attitude, Failure - Pearls of Advice, 485
Emergence of simple-cell receptive field properties by learning a sparse code for natural images, 486
Emergent properties of networks of biological signaling pathways, 487
Emergent Properties of the Optic Tectum Revealed by Population Analysis of Direction and Orientation Selectivity, 488
Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome, 489
Encoding Through Patterns: Regression Tree^^e2^^80^^93Based Neuronal Population Models, 490
Encryption is less secure than we thought, 491
Endogenous angiotensin II facilitates GABAergic neurotransmission afferent to the Na+-responsive neurons of the rat median preoptic nucleus., 492
Energy-efficient encoding by shifting spikes in neocortical neurons, 493
Engineering a memory with LTD and LTP, 494, 495
Engineering of weak helper interactions for high-efficiency FRET probes, 496
Entering Mentoring: A Seminar to Train a New Generation of Scientists, 497
Entopsis , 498
Environmentally induced epigenetic transgenerational inheritance of disease susceptibility, 499
Environmentally Induced Transgenerational Epigenetic Reprogramming of Primordial Germ Cells and the Subsequent Germ Line, 500
EosFP, 501
EPFL, 502
ephrins, 503
Epialleles in plant evolution, 504
Epigenetic and epigenomic variation in Arabidopsis thaliana, 505
Epigenetic inheritance of a cocaine-resistance phenotype, 506
Epigenetic mechanisms underlying learning and the inheritance of learned behaviors, 507
Epigenetic memory: the Lamarckian brain, 508
Epigenetic Transmission of the Impact of Early Stress Across Generations, 509
EPON, 510
Equating information-theoretic and likelihood-based methods for neural dimensionality reduction, 511
Ersatz, 512
Ethical reproducibility: towards transparent reporting in biomedical research, 513
Evaluation of Excess Significance Bias in Animal Studies of Neurological Diseases, 514
Evidence for a computational distinction between proximal and distal neuronal inhibition, 515
Evidence for a computational distinction between proximal and distal neuronal inhibition., 516
Evidence for Hubs in Human Functional Brain Networks, 517
Evolution Heresy? Epigenetics Underlies Heritable Plant Traits, 518
evolutionary correlations, 519
Evolutionary origins of the avian brain, 520
excellent set of notes, 521
Excitatory and inhibitory interactions in localized populations of model neurons, 522
Exotic optics: Metamaterial world, 523, 524
Expansion microscopy, 525
Experimental evidence needed to demonstrate inter- and trans-generational effects of ancestral experiences in mammals, 526
Explicit memory creation during sleep demonstrates a causal role of place cells in navigation, 527
eyeborg, 528
fairhall lab, 529
False-Positive Psychology: Undisclosed Flexibility in Data Collection and Analysis Allows Presenting Anything as Significant, 530
Fast two-layer two-photon imaging of neuronal cell populations using an electrically tunable lens, 531
Fast-conducting mechanoreceptors contribute to withdrawal behavior in normal and nerve injured rats, 532
FD NeuroTechnologies, 533
figure 2 of Packer, 2012, 534
figure 2 of Rickgauer, 2009, 535
figure 3b, 536
figure 4h, 537
Findings Regarding The Market Events Of May 6, 2010, 538
First GWAS hits for cognitive ability, 539
Flash Crash: Could it happen again?, 540
Flexible Control of Mutual Inhibition: A Neural Model of Two-Interval Discrimination, 541
Flexible control of mutual inhibition: a neural model of two-interval discrimination, 542
flickr page, 543
Flows of Research Manuscripts Among Scientific Journals Reveal Hidden Submission Patterns, 544
fMRI response to blue light delivery in the na^^c3^^afve brain: Implications for combined optogenetic fMRI studies, 545
foc.us, 546
Food Restriction Increases Glutamate Receptor-Mediated Burst Firing of Dopamine Neurons, 547
For Best Results, Forget the Bonus, 548
Fostering the Independence of New Investigators in Biomedical Research. - Where Are We Now?, 549
Freebase, 550
freely moving dual photostimulation and imaging, 551, 552, 553
From another angle: Differences in cortical coding between fine and coarse discrimination of orientation., 554
From circuits to behavior: a bridge too far?, 555
From circuits to behaviour in the amygdala, 556
frontiers neuro blog, 557
Functional imaging in the zebrafish retinotectal system using RGECO, 558
Functional labeling of neurons and their projections using the synthetic activity^^e2^^80^^93dependent promoter E-SARE, 559
functional magnetic resonance imaging, 560
functional near-infrared spectroscopy, 561
Functional patterned multiphoton excitation deep inside scattering tissue, 562
Fundamental Limits to Moore’s Law, 563
Funding grant proposals for scientific research: retrospective analysis of scores by members of grant review panel, 564
G-protein coupled receptors, 565
Gain Control Network Conditions in Early Sensory Coding, 566
galvos, 567
Gary Marcus, 568
Gatsby, 569
Gaurav Venkataraman, 570
GCaMP, 571, 572
GCaMP7, 573
GDF11, 574
Geiger counters, 575
Genetic and Pharmacological Evidence for a Novel, Intermediate Phase of Long-Term Potentiation Suppressed by Calcineurin, 576
Genetic Control of Nociceptive Sensory Neuron Development and Pain Behavior pm Grantome, 577
Genetically-encoded Voltage Indicators, 578
genome-engineering.org, 579
git, 580
Global Alliance for Genomics and Health, 581, 582
Global Epigenomic Reconfiguration During Mammalian Brain Development, 583
Google Anti-Aging Startup, Calico, Snags Big Names: Barron, Botstein, 584
Google Compute Engine, 585
Google Glass, 586
Google Research, 587
Google Scholar, 588, 589
Google technical report, 590
Google Trends, 591
Google^^e2^^80^^99s Grand Plan to Make Your Brain Irrelevant, 592
Gordon Research Conferences, 593
GPU, 594
Graduate school: the movie, 595
Grantome, 596
Graphene for Terahertz Applications, 597, 598
Graur and co. argued was what went wrong with the ENCODE project, 599
great overview article, 600
GRIN, 601
groupthink, 602
Guide to research techniques in neuroscience, 603
GWAS, 604
Hadly Lab, 605
Hanchuan Peng, 606
Handbook of Basal Ganglia Structure and Function, 607
Handbook of Biological Statistics, 608
Hang Lu, 609
Hang Lu Lab, 610
Haruhiko Bito, 611
Hausser, 612, 613
HDF, 614
Head-mountable high speed camera for optical neural recording, 615
Health Level 7, 616
Hebbian synapses: biophysical mechanisms and algorithms, 617
Helmchen Lab, 618
Henry Markham, 619
Heritable gene targeting in the mouse and rat using a CRISPR-Cas system, 620
High-resolution reconstruction of the beating zebrafish heart, 621
High-resolution whole-brain staining for electron microscopic circuit reconstruction, 622
High-speed recording of neural spikes in awake mice and flies with a fluorescent voltage sensor, 623
Highly nonrandom features of synaptic connectivity in local cortical circuits, 624
HoloLens, 625
How (not) to get a job..., 626
How does she do it?, 627
How learning can guide evolution., 628
How Many Scientists Fabricate and Falsify Research? A Systematic Review and Meta-Analysis of Survey Data, 629
How reliable is peer review? An examination of operating grant proposals simultaneously submitted to two similar peer review systems, 630
How to Be a Good Graduate Student – Succeeding in Graduate School, 631
How to Become a Successful Scientist, 632
How To Choose a Good Scientific Problem, 633
How to choose a thesis advisor?, 634
How to succeed in science: a concise guide for young biomedical scientists. Part I: taking the plunge, 635
How to succeed in science: a concise guide for young biomedical scientists. Part II: making discoveries, 636
htlatex, 637
http://bigwww.epfl.ch/thevenaz/turboreg/, 638, 639
http://dspcad-www.iacs.umd.edu/bcnm/index.html, 640
http://en.wikipedia.org/wiki/2010˙Flash˙Crash, 641
http://stnava.github.io/ANTs/, 642
http://uemweb.biomed.cas.cz/tpp/, 643
https://biaflows.neubias.org, 644, 645
https://bitbucket.org/adamshch/naomi˙sim/src/master/, 646, 647
https://cellprofiler.org, 648, 649
https://cran.r-project.org/web/packages/scalpel/index.html, 650, 651
https://cytomine.be, 652, 653
https://elastix.lumc.nl, 654
https://figshare.com/articles/dataset/Data/19193792, 655
https://github.com/adamshch/GraFT-analysis, 656
https://github.com/adamshch/SEUDO, 657
https://github.com/alexklibisz/deep-calcium, 658
https://github.com/AllenInstitute/deepinterpolation, 659, 660, 661, 662
https://github.com/amitani/onlineMotionCorrection, 663
https://github.com/bahanonu/ciatah, 664, 665, 666, 667, 668, 669
https://github.com/bahanonu/imaging˙tools, 670, 671
https://github.com/baidatong/NeuroSeg, 672, 673
https://github.com/BoHuangLab/Transfer-Learning-Denoising/, 674
https://github.com/cabooster/DeepCAD-RT, 675, 676
https://github.com/cabooster/DeepCAD-RT/, 677
https://github.com/cabooster/SRDTrans, 678
https://github.com/CaliAli-PV/CaliAli, 679
https://github.com/comp-imaging-sci/MVG-CNN, 680, 681
https://github.com/datajoint/datajoint-matlab, 682, 683
https://github.com/DeniseCaiLab/minian, 684, 685
https://github.com/EKirschbaum/DISCo, 686
https://github.com/flatironinstitute/CaImAn, 687, 688, 689, 690, 691, 692, 693, 694
https://github.com/flatironinstitute/CaImAn-MATLAB, 695, 696
https://github.com/HelmchenLabSoftware/Cascade, 697, 698
https://github.com/hochbaumGroup/HNCcorr, 699, 700
https://github.com/ikinsella/locaNMF, 701, 702
https://github.com/jf-lab/cnmfe-reviewer, 703, 704
https://github.com/kushalkolar/MESmerize, 705, 706
https://github.com/Leveltlab/SpectralSegmentation, 707, 708, 709
https://github.com/LieberInstitute/CaPTure, 710, 711, 712
https://github.com/losonczylab/sima, 713, 714
https://github.com/mouseland/cellpose, 715, 716
https://github.com/nel-lab/FIOLA, 717, 718
https://github.com/NeurodataWithoutBorders, 719, 720
https://github.com/NICALab/BEAR, 721, 722
https://github.com/NoahDolev/Segment2P, 723
https://github.com/NTCColumbia/moco, 724, 725
https://github.com/orlandi/netcal, 726, 727
https://github.com/paninski-lab/funimag, 728, 729
https://github.com/phflot/flow˙registration, 730
https://github.com/porteralab/EZcalcium, 731, 732
https://github.com/rmcassidy/FIBSI˙program, 733, 734
https://github.com/rochefort-lab/fissa, 735, 736
https://github.com/ryhattori/PatchWarp, 737, 738
https://github.com/samuroi/SamuROI, 739, 740
https://github.com/schnitzer-lab/EXTRACT-public, 741, 742
https://github.com/SharifAmit/4SM, 743
https://github.com/SilverLabUCL/SilverLab-Microscope, 744
https://github.com/simonsfoundation/NoRMCorre, 745, 746
https://github.com/sshen8/acsat, 747
https://github.com/StephanieRey/ABLE, 748, 749
https://github.com/tzklab/carignan, 750
https://github.com/Weiyi-Liu-Unique/FIFER, 751
https://github.com/XZH-James/NeuroSeg2, 752
https://github.com/YijunBao/Shallow-UNet-Neuron-Segmentation˙SUNS, 753
https://github.com/yuanlong-o/Deep˙widefield˙cal˙inferece, 754
https://github.com/zebrain-lab/Toolbox-Romano-et-al, 755, 756
https://github.com/zhe-ch/ACTEV, 757
https://github.com/zhoupc/CNMF˙E, 758, 759
https://github.com/zhoupc/OASIS˙matlab, 760, 761
https://github.com/zivlab/CellReg, 762, 763
https://gitlab.com/anflores/axial˙motion˙correction, 764
https://gitlab.com/cossartlab/deepcinac, 765
https://gitlab.iit.it/fellin-public/astra, 766
https://gitlab.iit.it/fellin-public/cite-on, 767
https://icy.bioimageanalysis.org/plugin/elastic-motion-correction-concatenation-emc2-of-tracks/, 768
https://openaccess.thecvf.com/content/WACV2023/supplemental/Cho˙Robust˙and˙Efficient˙WACV˙2023˙supplemental.zip, 769
https://openbis.ch, 770, 771
https://www.bu.edu/hanlab/files/2016/02/pfgc.zip, 772
https://www.knime.com, 773, 774
https://www.openmicroscopy.org, 775, 776
Human Brain Project, 777, 778
human brain project, 779
Human brain volume, 780
Human Connectome Project, 781
Human-level control through deep reinforcement learning, 782
Huntington disease arises from a combinatory toxicity of polyglutamine and copper binding, 783
I listen to color, 784
IBM Scientists Show Blueprints for Brain-like Computing, 785
Identification of a splice variant of mouse TRPA1 that regulates TRPA1 activity, 786
Identification of Spinal Circuits Transmitting and Gating Mechanical Pain, 787
iGluSnFR, 788, 789
Igor Markov, 790
Imaging calcium microdomains within entire astrocyte territories and endfeet with GCaMPs expressed using adeno-associated viruses, 791
Imaging Circulating Tumor Cells in Freely Moving Awake Small Animals Using a Miniaturized Intravital Microscope, 792
Imaging electrical activity of neurons with metamaterial nanosensors, 793
Imaging neural spiking in brain tissue using FRET-opsin protein voltage sensors, 794
Imaging of Tau Pathology in a Tauopathy Mouse Model and in Alzheimer Patients Compared to Normal Controls, 795
Imaging the Awake Visual Cortex with a Genetically Encoded Voltage Indicator, 796
Imaging zebrafish neural circuitry from whole brain to synapse, 797
immediate early gene, 798
Impact Factor Distortions, 799
Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice, 800
In Defense of Academic Writing, 801
In focus: molecular and cell biology research in China, 802
In it to win it: Pathway to scientific independence by the NIH K99 award, 803
In vivo Calcium Imaging to Illuminate Neurocircuit Activity Dynamics Underlying Naturalistic Behavior, 804
In vivo evidence that retinal bipolar cells generate spikes modulated by light, 805
In vivo optogenetic identification and manipulation of GABAergic interneuron subtypes, 806
In vivo robotics: the automation of neuroscience and other intact-system biological fields, 807
In vivo synaptic recovery following optogenetic hyperstimulation, 808
In vivo time-gated fluorescence imaging with biodegradable luminescent porous silicon nanoparticles, 809
in-situ, 810
Independent circuits in the basal ganglia for the evaluation and selection of actions, 811
Inflammatory Pain Promotes Increased Opioid Self-Administration: Role of Dysregulated Ventral Tegmental Area mu Opioid Receptors, 812
Influence of dendritic structure on firing pattern in model neocortical neurons, 813
Information and Efficiency in the Nervous System–A Synthesis, 814
Information and Efficiency in the Nervous System^^e2^^80^^94A Synthesis, 815
Inorganic materials: Intuition weaved into computation, 816
Inscopix, 817
Inspiration Mars Foundation, 818
Integrated information theory , 819
Integration of GABAergic Interneurons into Cortical Cell Assemblies: Lessons from Embryos and Adults, 820
Intel Galileo, 821
Intel PRO/1000 GT Desktop Adapter, 822
Interaction of FUS and HDAC1 regulates DNA damage response and repair in neurons, 823
Interesting (Computational) Neuroscience Papers, 824, 825
Internally generated cell assembly sequences in the rat hippocampus, 826
International Neuroinformatics Coordinating Facility, 827
Intrinsic and network rhythmogenesis in a reduced Traub model for CA3 neurons, 828
Investigating the role of ^^ef^^ac^^81ring-rate normalization and dimensionality reduction in brain-machine interface robustness, 829
Ion-channel defects and aberrant excitability in myotonia and periodic paralysis, 830
IPTG, 831
Is there a common molecular pathway for addiction?, 832
It’s the Effect Size, Stupid What effect size is and why it is important, 833, 834
JAABA: interactive machine learning for automatic annotation of animal behavior, 835
Jan Grundemann, 836
Janelia, 837
Janelia Farm, 838
Jasanoff lab, 839
JAX, 840, 841
JAX Allen Mice, 842
Jerome Lecoq, 843
Jin Hyung Lee, 844
Jonathan Victor, 845
Jones Parker, 846
Journal of Neuroscience, 847, 848
Kainate Receptors Mediate Signaling in Both Transient and Sustained OFF Bipolar Cell Pathways in Mouse Retina, 849
Kang Shen, 850
Karel Svoboda, 851
Kay Tye, 852
Keller, 2013, 853
Kevin Briggman, 854
Koga, Neuron 2015, 855
Kunle Olukotun, 856
Labrigger, 857, 858, 859, 860, 861
labrigger, 862
Lamarck revisited: epigenetic inheritance of ancestral odor fear conditioning, 863
Lamarckism, 864
Laminar optical tomography: demonstration of millimeter-scale depth-resolved imaging in turbid media, 865
Laminar optical tomography: high-resolution 3D functional imaging of superficial tissues, 866
large scale imaging and other papers, 867, 868
Large-scale navigational map in a mammal, 869
Large-scale, high-density (up to 512 channels) recording of local circuits in behaving animals, 870
Larsch, 2013, 871
Latent Dirichlet allocation, 872
latex boilerplate, 873
layman’s report, 874
Lazy Load, 875
Learn X in Y minutes, 876
Learning representations by back-propagating errors, 877
leave a comment below, 878, 879
Lee Rubin, 880
Lee, 2014, 881
Lee, J Neuro 2015, 882
legal battle over CRISPR, 883
Lessons learned from the fate of AstraZeneca’s drug pipeline: a five-dimensional framework, 884
Levene lab, 885
light sheet microscopes, 886
Light trials for human blindness, 887
Light-evoked Somatosensory Perception of Transgenic Rats That Express Channelrhodopsin-2 in Dorsal Root Ganglion Cells, 888
light-sheet microscopy, 889
Lighting The Brain - Karl Deisseroth and the optogenetics breakthrough., 890
Limiting the Impact of the Impact Factor, 891
limits of computation, 892
Limits on fundamental limits to computation, 893
Lin Tian, 894
Lineage-specific laminar organization of cortical GABAergic interneurons, 895
Linguistic relativity, 896
link, 897, 898
liposomes, 899
Liu Lab, 900
locus coeruleus, 901
long term depression, 902
Looger, 903
Loren Looger, 904
LTD, 905
LTP, 906
M16 - Optogenetic inhibition of specific populations of sensory neurons mediating bladder nociception, 907
Machine Learning: The Art and Science of Algorithms that Make Sense of Data, 908
Magic Leap, 909
Magneto-fluorescent core-shell supernanoparticles, 910
Mainak Chowdhury, 911
Making data sharing work: The FCP/INDI experience, 912
manganese, 913
Manipulating circadian clock neuron firing rate resets molecular circadian rhythms and behavior, 914
Mapping Neuronal Diversity One Cell at a Time, 915
MapReduce, 916
Marblestone and others, 917
Marder, 918
Mark Schnitzer, 919
Mark Schnitzer’s lab, 920, 921
Markram, 922
marmoset’s as model organisms, 923
Marvin, 2013, 924
Mechanisms of epigenetic memory and addiction, 925
medial forebrain bundle, 926
MeSH, 927
Metabotropic glutamate receptors in median preoptic neurons modulate neuronal excitability and glutamatergic and GABAergic inputs from the subfornical organ., 928
metaio, 929
Michael Fee, 930, 931
microarray analysis, 932
Microfluidic Device for Single-Cell Analysis, 933
Microfluidic Platforms for Single-Cell Analysis, 934
Microfluidics for in vivo imaging of neuronal and behavioral activity in Caenorhabditis elegans, 935
Microfluidics for single cell analysis, 936
Microfluidics for the analysis of behavior, nerve regeneration, and neural cell biology in C. elegans, 937
Microglia Disrupt Mesolimbic Reward Circuitry in Chronic Pain, 938
Micropatterned substrates coated with neuronal adhesion molecules for high-content study of synapse formation, 939
MicroscopyU, 940
Microsoft HoloLens, 941
miniature microscopes, 942, 943
Misha Ahrens, 944
MIT, 945
MIT Comp Biology, 946, 947
MIT Technology Review, 948, 949
Mo Papers Mo Problems, 950, 951, 952
Modha, 953
molecular imprinting, 954
Molecular mechanisms for the inheritance of acquired characteristics^^e2^^80^^94exosomes, microRNA shuttling, fear and stress: Lamarck resurrected?, 955
Molecular-Level Functional Magnetic Resonance Imaging of Dopaminergic Signaling, 956
Moneyball, 957
Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons., 958
More Perfect Than We Imagined: A Physicist’s View, 959
Motion illusions as optimal percepts., 960
Motor Cortex Feedback Influences Sensory Processing by Modulating Network State, 961
Mouse brain imaging using photoacoustic computed tomography, 962
Mouse Connectome Project, 963
Mouse Imaging Centre, 964, 965
Mrgprd-Expressing Polymodal Nociceptive Neurons Innervate Most Known Classes of Substantia Gelatinosa Neurons, 966
MRI contrast agents, 967
much like the PS3 was, 968
Mud Sticks: On the Alleged Falsification of Mendel’s Data, 969
Multi-core processors, 970
Multi-task connectivity reveals flexible hubs for adaptive task control, 971
multi-threading, 972
multiple organ failure, 973
Multiplicative computation in a visual neuron sensitive to looming, 974
Multipoint-Emitting Optical Fibers for Spatially Addressable In Vivo Optogenetics, 975
Munos On Big Companies and Small Ones, 976
Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders, 977
Mutso 2012, 978
my previous post on the subject, 979
Nachum Ulanovsky, 980
Nanoscale-Targeted Patch-Clamp Recordings of Functional Presynaptic Ion Channels, 981
Nat Clarke, 982
Natural Neural Projection Dynamics Underlying Social Behavior, 983
Natural Neural Projection Dynamics Underlying Social Behavior (figure 8), 984
Nature article, 985
Nature Biotechnology, 986, 987
Nature Chemistry, 988, 989
Nature Communications, 990, 991
Nature Medicine, 992
Nature Method, 993, 994
Nature Nanotechnology, 995, 996
Nature Neuroscience, 997, 998
Nature promotes read-only sharing by subscribers, 999
Nature publishes overwhelmingly proven “NEW AMAZING FINDING”....because optogenetics, 1000
Nature Reviews Neuroscience, 1001, 1002
netfabb, 1003
netrin, 1004
Network deconvolution as a general method to distinguish direct dependencies in networks, 1005
Network link prediction by global silencing of indirect correlations, 1006
Network motifs: simple building blocks of complex networks, 1007
Neural circuits and behaviour of Drosophila, 1008
Neural Computation, 1009, 1010, 1011
Neural networks and brain function, 1012
Neural networks and physical systems with emergent collective computational abilities, 1013
Neural Representation and the Cortical Code, 1014
Neural Representation of a Target Auditory Memory in a Cortico-Basal Ganglia Pathway, 1015
Neural Syntax: Cell Assemblies, Synapsembles, and Readers, 1016
NeuralMapper, 1017, 1018
Neurobasal, 1019
Neurocommons, 1020, 1021, 1022
Neurodata Without Borders, 1023
Neurodudes, 1024, 1025, 1026
NeuroElectro, 1027
Neuroendocrine Control of Drosophila Larval Light Preference, 1028
neurogenesis, 1029
neuromorphic engineering, 1030
neuromuscular junctions, 1031
Neuron, 1032, 1033
Neuronal circuits that regulate feeding behavior and metabolism, 1034
Neuronal correlates of parametric working memory in the prefrontal cortex, 1035
Neuronal generation of the leech swimming movement, 1036, 1037
Neuronal generation of the leech swimming movement., 1038
Neuronal networks of the hippocampus, 1039
Neurons with graded response have collective computational properties like those of two-state neurons, 1040
Neuropeptide signaling remodels chemosensory circuit composition in Caenorhabditis elegans, 1041
Neuroscience and education: myths and messages, 1042
NeuroScience Associates, 1043
Neuroscience Fiction, 1044
neuroscience imaging analysis tools, 1045, 1046
Neuroscience in the 21st Century, 1047
Neuroscience thinks big (and collaboratively), 1048
Neuroscience: Brain projects need stronger foundation, 1049
Neuroscience: Map the other brain, 1050
Neuroscience: Solving the brain, 1051
NeuroSky, 1052
NeuWrite West, 1053
new article, 1054
New details on google anti-aging., 1055
new neuroscience tools from janelia, 1056
New Optogenetic System Shines a Light on Pain-Sensing Neurons, 1057
news article, 1058
NewScientist, 1059
NewScientist: 2015 before it happens, 1060
Newsome, 1061
Newt Gingrich: Double the N.I.H. Budget, 1062
NIH and Biomedical ^^e2^^80^^98Big Data^^e2^^80^^99, 1063
NIH Big Data to Knowledge (BD2K) initiative, 1064, 1065, 1066
NIH BRAIN awards, 1067
NIH program on Rigor and Reproducibility, 1068
NIH RePORT, 1069
NIH RePORTER, 1070
NIH review committees, 1071
NIH Working Group on Data and Informatics, 1072, 1073
NIH’s RePORT site, 1074
No, You’re Not an Impostor, 1075
Nobel Prize Browser, 1076, 1077
Nobie Redmon, 1078, 1079, 1080
Nociception and pain: lessons from optogenetics, 1081
Noise characteristics and prior expectations in human visual speed perception, 1082
Non-redundant odor coding by sister mitral cells revealed by light addressable glomeruli in the mouse, 1083
nonribosomal peptide synthesis, 1084
Norbinaltorphimine, 1085
Nuclear calcium signaling in the regulation of brain function, 1086
Nuclear Lamin-A Scales with Tissue Stiffness and Enhances Matrix-Directed Differentiation, 1087
Numenta, 1088
NYTimes article here, 1089
Oath of the Horatii, 1090
Objet24, 1091
octopuses, 1092
ocular dominance columns, 1093
Oculus VR, 1094, 1095
Odor Discrimination in Drosophila: From Neural Population Codes to Behavior, 1096
On brain activity mapping: insights and lessons from Brain Decoding Project to map memory patterns in the hippocampusOn brain activity mapping: insights and lessons from Brain Decoding Project to map memory patterns in the hippocampus, 1097
On the reproducibility of science: unique identification of research resources in the biomedical literature, 1098
On writing well, 1099
One-Step Generation of Mice Carrying Reporter and Conditional Alleles by CRISPR/Cas-Mediated Genome Engineering, 1100, 1101
Open Neuroscience, 1102, 1103, 1104
Open Optogenetics, 1105, 1106, 1107
Open Wetware, 1108, 1109, 1110
OpenCV, 1111
OpenOpto, 1112
optical, 1113
Optical Alignment, 1114
Optical control of mammalian endogenous transcription and epigenetic states, 1115
Optical Control of Muscle Function by Transplantation of Stem Cell^^e2^^80^^93Derived Motor Neurons in Mice, 1116
Optical fibers for high-resolution in vivo microendoscopic fluorescence imaging, 1117
optical orthogonal frequency division multiplexing, 1118
optical resonators, 1119
optical RFID, 1120
Optical tomography, 1121
Optics Letters, 1122, 1123
Optimizing Sound Features for Cortical Neurons, 1124
optogenetic, 1125
Optogenetic Control of Targeted Peripheral Axons in Freely Moving Animals, 1126, 1127
Optogenetic identification of striatal projection neuron subtypes during in vivo recordings, 1128
Optogenetic Recruitment of Dorsal Raphe Serotonergic Neurons Acutely Decreases Mechanosensory Responsivity in Behaving Mice, 1129, 1130
Optogenetic Regeneration, 1131
Optogenetic stimulation of a hippocampal engram activates fear memory recall., 1132
Optogenetically Induced Behavioral and Functional Network Changes in Primates, 1133
optogenetics, 1134
optogenetics and pain, 1135, 1136
optogenetics and pain, continued, 1137
Optogenetics at a crossroads?, 1138
optogenetics identification of specific cell types, 1139
Optogenetics Resource Center, 1140
optogenetics wiki, 1141
Orbitofrontal and striatal circuits dynamically encode the shift between goal-directed and habitual actions, 1142
ORCID identifier, 1143
Overexpression of calcineurin in mouse causes sudden cardiac death associated with decreased density of K+ channels, 1144
Pacific Rim, 1145
Pain: from new perspectives to novel treatments, 1146
Palantir, 1147
Panasonic and Imec joint project, 1148
parabiosis, 1149
Parallel GPU Implementation of Iterative PCA Algorithms, 1150
Parametric Functional Maps of Visual Inputs to the Tectum, 1151
Parental olfactory experience influences behavior and neural structure in subsequent generations, 1152
Paternal Stress Exposure Alters Sperm MicroRNA Content and Reprograms Offspring HPA Stress Axis Regulation, 1153
Paternally Induced Transgenerational Environmental Reprogramming of Metabolic Gene Expression in Mammals, 1154
Paul G. Allen to Give 100 Million to Create Cell Science Institute, 1155
Pay Enough or Don’t Pay at All, 1156
Peer review for improving the quality of grant applications, 1157
PEGylation, 1158
Percentile Ranking and Citation Impact of a Large Cohort of NHLBI-Funded Cardiovascular R01 Grants, 1159
Permanent Genetic Access to Transiently Active Neurons via TRAP: Targeted Recombination in Active Populations, 1160
persistent homology, 1161
Pervasive Parallelism Lab, 1162, 1163
Pfizer’s R&D Productivity, 1164
pharma projects, 1165
PhD Students: Should You Switch Labs?, 1166
PhD survival guide, 1167
Phi: A Voyage from the Brain to the Soul , 1168
Philosophers are Mortal: Inferring the Truth of Unseen Facts, 1169
phonons, 1170
photoacoustic imaging, 1171
Photoactivatable Neuropeptides for Spatiotemporally Precise Delivery of Opioids in Neural Tissue, 1172
Photonics West 2015, 1173
Physical Review Letters, 1174
physiological optogenetics, 1175
PicnicHealth, 1176
PicnicHealth Stores Your Medical Records In One Place And Delivers It To Your Doctor, 1177
Pilot safety study of iPSC-based intervention for wet-type AMD, 1178
Pittsburgh, 1179
Place Cells, Grid Cells, and the Brain’s Spatial Representation System, 1180
placozoa, 1181
Plasmonic gold mushroom arrays with refractive index sensing figures of merit approaching the theoretical limit, 1182
Playing Atari with Deep Reinforcement Learning, 1183, 1184
Playing with Fire: Linking Intelligence to Our Genetics, 1185
PLoS Biology, 1186, 1187
PLOS Computational Biology, 1188, 1189
PMT, 1190
pointers, 1191
Polbase, 1192
Politics and the English Language, 1193, 1194
PON systems, 1195
Poor Productivity As A Self-inflicted Injury: Who^^e2^^80^^99s Missing The Most Toes, And Why, 1196
Population Coding and the Labeling Problem: Extrinsic Versus Intrinsic Representations, 1197
positron emission tomography, 1198
Postdoc payday: Salaries for fellows are on the up, 1199
potentiation, 1200
Power failure: why small sample size undermines the reliability of neuroscience, 1201
Prediction and validation of the distinct dynamics of transient and sustained ERK activation., 1202
Predictive coding: a fresh view of inhibition in the retina, 1203
Principles Governing the Operation of Synaptic Inhibition in Dendrites, 1204
Prion-like transmission of neuronal huntingtin aggregates to phagocytic glia in the Drosophila brain, 1205
Prion-like transmission of protein aggregates in neurodegenerative diseases, 1206
Probabilistic brains: knowns and unknowns, 1207
Probing perceptual decisions in rodents, 1208
Productivity Metrics and Peer Review Scores, 1209
Productivity versus age, 1210
Prolonged dopamine signalling in striatum signals proximity and value of distant rewards, 1211
promoters, 1212
Prostaglandin signaling suppresses beneficial microglial function in Alzheimer^^e2^^80^^99s disease models, 1213
protein tags, 1214
Psychology Journal Bans Significance Testing, 1215
PTC Creo, 1216, 1217
pubchase, 1218
Pubmed, 1219, 1220
pumps, 1221
Puzzle Imaging: Using Large-scale Dimensionality Reduction Algorithms for Localization, 1222
Q3 - Characterization of optogenetic activation of non-peptidergic C-fibers, 1223
quantum, 1224
Quiet debut for the double helix, 1225
R01 grants, 1226
R&D productivity: on the comeback trail, 1227
Randal Burns, 1228
Rank injustice, 1229
Rapid 3D light-sheet microscopy with a tunable lens, 1230
Rapid evaluation of a protein-based voltage probe using a field-induced membrane potential change, 1231
Rapid local synchronization of action potentials: Toward computation with coupled integrate-and-fire neurons, 1232
Rapid optical control of nociception with an ion-channel photoswitch, 1233
ReaChR, 1234
ReaChR: a red-shifted variant of channelrhodopsin enables deep transcranial optogenetic excitation, 1235
Read before you cite, 1236
recent comments Elon Musk, 1237
recently posted, 1238
Recovery from slow inactivation in K+ channels is controlled by water molecules, 1239
Regularization theory and neural networks architectures., 1240
Reinforcement, Reward, and Intrinsic Motivation: A Meta-Analysis, 1241
Relationship between intracortical electrode design and chronic recording function, 1242
Remote Optogenetic Activation and Sensitization of Pain Pathways in Freely Moving Mice, 1243
Report on Enhancing Peer Review at NIH Implementation Plan, 1244
Representation of Three-Dimensional Space in the Hippocampus of Flying Bats, 1245
ResearcherID, 1246
resonant cavities, 1247
Resources for surviving and thriving in graduate school (with a focus on the sciences) , 1248
Restoring Systemic GDF11 Levels Reverses Age-Related Dysfunction in Mouse Skeletal Muscle, 1249
Restricted and Regulated Overexpression Reveals Calcineurin as a Key Component in the Transition from Short-Term to Long-Term Memory, 1250
review 1, 1251
review 2, 1252
Reward Learning Requires Activity of Matrix Metalloproteinase-9 in the Central Amygdala, 1253
RGS9-2–controlled adaptations in the striatum determine the onset of action and efficacy of antidepressants in neuropathic pain states, 1254
rhesus monkey, 1255
Robotics and the Lessons of Cyberlaw, 1256
Robust multicellular computing using genetically encoded NOR gates and chemical ‘wires’, 1257
rodents into virtual reality, 1258
Ron Kopito, 1259
Rosetta begins its Comet Tale, 1260
Rosetta Brains: A Strategy for Molecularly-Annotated Connectomics, 1261
Ryan Lewis, 1262
Sabatini lab, 1263
Sample Size and Precision in NIH Peer Review, 1264
Sanes, 1265
SASER, 1266
SBIRP, 1267
Scalable Brain Atlas (3D models), 1268
Scanless two-photon excitation of channelrhodopsin-2, 1269, 1270
scanning electron microscope, 1271
Schema, 1272
Scherer, 1273
Schnitzer, 1274
Schnitzer Lab, 1275
Schnitzer lab, 1276
Science, 1277
Science Isn^^e2^^80^^99t Broken, 1278
Science Lab, 1279
Science’s perspective piece, 1280
Sciencescape, 1281
Sciencescape (archive link), 1282
Sciencescape (original, dead), 1283
Sciencescape (updated link), 1284
Scientific American, 1285, 1286
Scientific method: Defend the integrity of physics, 1287
Scientific method: Statistical errors, 1288, 1289
Scientists sound alarm over DNA editing of human embryos, 1290
SciTrends, 1291, 1292, 1293
SDCube and hybrid data storage, 1294, 1295
Sebastian Seung, 1296
Sebastian Seung TED talk, 1297
Sebastian Seung’s Quest to Map the Human Brain, 1298
seed, 1299
SeeDB, 1300, 1301
Seeing the Natural World With a Physicist^^e2^^80^^99s Lens, 1302
Segregation of object and background motion in the retina, 1303
Sejnowski, 1304
Self-propagation of pathogenic protein aggregates in neurodegenerative diseases, 1305
Serial optical coherence scanner for large-scale brain imaging at microscopic resolution, 1306
Serotonin and the Neuropeptide PDF Initiate and Extend Opposing Behavioral States in C. elegans, 1307
Seung, 1308
SGD, 1309
Shank3 mutant mice display autistic-like behaviours and striatal dysfunction, 1310
Shannon’s Bandwagon essay, 1311
Shapiro, 2010, 1312
shards, 1313
Sheer Economics: How We Got in This Fix, 1314
Shen, 1315
SHH, 1316
Shocking Revelations and Saccharin Sweetness in the Study of Drosophila Olfactory Memory, 1317
Significance testing, 1318
similar project at Janelia for Drosophila, 1319
Simons Collaboration on the Global Brain: Scientific Mission of the Collaboration, 1320
Simplicial Models and Topological Inference in Biological Systems, 1321
Simultaneous all-optical manipulation and recording of neural circuit activity with cellular resolution in vivo, 1322, 1323
Simultaneous cellular-resolution optical perturbation and imaging of place cell firing fields, 1324
Simultaneous generation and germline transmission of multiple gene mutations in rat using CRISPR-Cas systems, 1325
Simultaneous PET-MRI reveals brain function in activated and resting state on metabolic, hemodynamic and multiple temporal scales, 1326
Simultaneous whole-animal 3D imaging of neuronal activity using light-field microscopy, 1327
Singapore’s salad days are over, 1328
Single-cell microfluidic impedance cytometry: a review, 1329
Single-shot compressed ultrafast photography at one hundred billion frames per second, 1330, 1331
singularity, 1332
Skin ^^ce^^b2-Endorphin Mediates Addiction to UV Light, 1333
SmartScope, 1334
smFP, 1335
Smith, 1336
Social Science Genetic Association Consortium, 1337
Society for Neuroscience Conference, 1338
Soft tissue preservation in a fossil marine lizard with a bilobed tail fin, 1339
SOLIDWORKS, 1340, 1341
Sorger lab, 1342
source, 1343
SpaceX, 1344
spaghetti monster, 1345
sparse coding, 1346, 1347, 1348
Sparse Coding Models Can Exhibit Decreasing Sparseness while Learning Sparse Codes for Natural Images, 1349
Sparse coding of sensory inputs., 1350
Spatial cognition in bats and rats: from sensory acquisition to multiscale maps and navigation, 1351
spatial light modulators, 1352, 1353
spatial navigation in bats, 1354
Spatially Selective Holographic Photoactivation and Functional Fluorescence Imaging in Freely Behaving Mice with a Fiberscope, 1355, 1356
Spatiotemporal Control of Opioid Signaling and Behavior, 1357
Spike train metrics, 1358
Spikes: Exploring the Neural Code (Computational Neuroscience), 1359
SPIM, 1360
Stanford, 1361
stanford neuroblog, 1362
Stanford news, 1363
Stanford University researchers found a cure for Alzheimer’s disease, 1364
Statistical analysis of the National Institutes of Health peer review system, 1365
Statistics for biologists, 1366, 1367
statistics: effect sizes, 1368
Steady or changing? Long-term monitoring of neuronal population activity, 1369
Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells., 1370
Stowers Institute for Medical Research, 1371
Stress in Biomedical Research: Six Impossible Things, 1372
Structure and Interpretation of Computer Programs, 1373
Structure-Guided Directed Evolution of Highly Selective P450-Based Magnetic Resonance Imaging Sensors for Dopamine and Serotonin, 1374
Structure-Guided Transformation of Channelrhodopsin into a Light-Activated Chloride Channel, 1375
substantia nigra, 1376
Superintelligence Paths, Dangers, Strategies, 1377
Superintelligence: Paths, Dangers, Strategies, 1378
supplemental info, 1379
Suppressing aberrant GluN3A expression rescues synaptic and behavioral impairments in Huntington’s disease models, 1380
Suppression of the morphine-induced rewarding effect in the rat with neuropathic pain: implication of the reduction in mu-opioid receptor functions in the ventral tegmental area, 1381
Suprachiasmatic stimulation phase shifts rodent circadian rhythms, 1382
survive exposure to the vacuum and radiation of space, 1383
Surviving a bioscience Ph.D., 1384
Swept confocally-aligned planar excitation (SCAPE) microscopy for high-speed volumetric imaging of behaving organisms, 1385
SwissTech Convention Center, 1386
Synaptic depression and cortical gain control, 1387
Synchronized oscillations in interneuron networks driven by metabotropic glutamate receptor activation, 1388
Syntactic Ngrams over Time, 1389
Synthesis of models for excitable membranes, synaptic transmission and neuromodulation using a common kinetic formalism, 1390
Synthesizing cognition in neuromorphic electronic systems, 1391
systems neuroscience, 1392
taking care with statistics, 1393
tamoxifen, 1394
Tank, 1395
Tank injustice and academic promotion, 1396
tardigrades, 1397
Targeted Ablation, Silencing, and Activation Establish Glycinergic Dorsal Horn Neurons as Key Components of a Spinal Gate for Pain and Itch, 1398
Targeted genome modification of crop plants using a CRISPR-Cas system, 1399
Targeted optogenetic stimulation and recording of neurons in vivo using cell-type-specific expression of Channelrhodopsin-2, 1400
Targeting neurons and photons for optogenetics, 1401
Taste Response Variability and Temporal Coding in the Nucleus of the Solitary Tract of the Rat, 1402
temporal exponential random graphical models, 1403
Temporal information transformed into a spatial code by a neural network with realistic properties, 1404
Temporally Precise Cell-Specific Coherence Develops in Corticostriatal Networks during Learning, 1405
Temporally precise in vivo control of intracellular signalling, 1406
Testing a Point Null Hypothesis: The Irreconcilability of P Values and Evidence, 1407
tetracycline, 1408
tetrodes, 1409
Thalamocortical oscillations in the sleeping and aroused brain, 1410
The ‘independent components’ of natural scenes are edge filters, 1411
The Abuse of Power: The Pervasive Fallacy of Power Calculations for Data Analysis, 1412
The Agony and the Ecstasy^^e2^^80^^94 The History and Meaning of the Journal Impact Factor, 1413
The AI Revolution: The Road to Superintelligence, 1414
The AI-Box Experiment, 1415
The Basic AI Drives, 1416
The benefits of brain mapping, 1417
The blue brain project, 1418
The BRAIN Initiative: developing technology to catalyse neuroscience discovery, 1419
the clever machine, 1420
The COMBREX Project: Design, Methodology, and Initial Results, 1421
The Coming Technological Singularity: How to Survive in the Post-Human Era, 1422
The computational brain, 1423
The correlation theory of brain function, 1424
The Diffraction Barrier in Optical Microscopy, 1425
The effects of FreeSurfer version, workstation type, and Macintosh operating system version on anatomical volume and cortical thickness measurements, 1426
The Elements of Style, 1427
The emergence of functional microcircuits in visual cortex, 1428
The Evolving Postdoctoral Experience, 1429
The Extraordinary Evolutionary History of the Reticuloendotheliosis Viruses, 1430
The First International Optogenetic Therapies for Vision Symposium, 1431
The Fundamental Physical Limits of Computation, 1432
The future of human cerebral cartography: a novel approach, 1433
The Future of Seawater Desalination: Energy, Technology, and the Environment, 1434
The Future of the Mind, 1435
The importance of stupidity in scientific research, 1436
The Laboratory Mouse, 1437
the Malinow lab, 1438
the Martian, 1439
The mismeasurement of science, 1440
The Misused Impact Factor, 1441
The monolith and the markets, 1442
The Moral Hazards and Legal Conundrums of Our Robot-Filled Future, 1443
The Most Dangerous Ideas In Science, 1444
The Mouse in Biomedical Research, 1445
The Mouse Nervous System, 1446, 1447
The necessity of animal models in pain research, 1448
The Need for Research Maps to Navigate Published Work and Inform Experiment Planning, 1449
The neural code between neocortical pyramidal neurons depends on neurotransmitter release probability, 1450
The neuron classification problem, 1451
The neuronal encoding of information in the brain, 1452
The Neuroterrain 3D Mouse Brain Atlas, 1453
the new clearing techniques as i’ve discussed previously, 1454
The New Fowler’s Modern English Usage, 1455
The office of the future: a unified approach to image-based modeling and spatially immersive displays, 1456
The origins and the future of microfluidics, 1457
The PhD journey: how to choose a good supervisor, 1458
The politics of publication, 1459
The Predictive Ability of Peer Review of Grant Proposals: The Case of Ecology and the US National Science Foundation, 1460
The Programmer’s Apprentice Project: A Research Overview, 1461
The Quest for Artificial Intelligence, 1462
The Rat Nervous System by George Paxinos, 1463
the return of the brain initiative notes, 1464, 1465
The rise of the professional master^^e2^^80^^99s degree: the answer to the postdoc/PhD bubble, 1466
The role of dendrites in auditory coincidence detection, 1467
The role of the suprachiasmatic nuclei in the generation of circadian rhythms in the golden hamster, Mesocricetus auratus, 1468
The search for novel analgesics: re-examining spinal cord circuits with new tools, 1469
The Singularity Is Near: When Humans Transcend Biology, 1470
The SwissTech Convention Center, a lab for conferences of the future, 1471
The top 100 papers, 1472
The Trouble With Brain Science, 1473
The warrior in the machine: neuroscience goes to war, 1474
Theoretical and computational neuroscience, 1475
Theoretical Neuroscience, 1476
Thermodynamic Cost of Reversible Computing, 1477
Thirst driving and suppressing signals encoded by distinct neural populations in the brain, 1478
this diagram, 1479
this figure, 1480
this new report gives a nice overview, 1481
Three Examples Of Applied and Computational Homology, 1482
Three-dimensional head-direction coding in the bat brain, 1483
three-photo imaging, 1484
Thy1, 1485
Tick-Tock, 1486
TIFF, 1487
Time crystals, 1488
Time-dependent systolic and diastolic function in mice overexpressing calcineurin, 1489
time-division multiplexing, 1490
TITLE, 1491
Tom Dean, 1492, 1493
Too Much Success for Recent Groundbreaking Epigenetic Experiments, 1494
Tools for Resolving Functional Activity and Connectivity within Intact Neural Circuits, 1495
Top 10 Innovations 2014, 1496
Topographic Representation of Numerosity in the Human Parietal Cortex, 1497
Topoisomerase inhibitors unsilence the dormant allele of Ube3a in neurons, 1498
Topoisomerases facilitate transcription of long genes linked to autism, 1499
Topological Pattern Recognition for Point Cloud Data, 1500
Topology for computing, 1501
Training-induced and electrically induced potentiation in the neocortex, 1502
Transcranial magnetic stimulation, 1503
Transgenerational Epigenetic Inheritance: Myths and Mechanisms, 1504
Transgenerational Epigenetic Inheritance: Prevalence, Mechanisms, and Implications for the Study of Heredity and Evolution, 1505
transgenerational epigenetics, 1506
Transgenic and knockout databases: Behavioral profiles of mouse mutants, 1507
Transgenic Mice for Intersectional Targeting of Neural Sensors and Effectors with High Specificity and Performance, 1508
transmission electron microscopy, 1509
Transmitting Pain and Itch Messages: A Contemporary View of the Spinal Cord Circuits that Generate Gate Control, 1510
Transplantation reveals regional differences in oligodendrocyte differentiation in the adult brain, 1511
TRAP, 1512
Treating brain disorders with neuromodulation, 1513
trend toward open hardware, 1514
Trends in the production of scientific data analysis resources, 1515
Tricyclic antidepressant, 1516
Troy Astorino, 1517
Tsien, 1518
Tunable Optics, 1519
Two-photon excitation in scattering media by spatiotemporally shaped beams and their application in optogenetic stimulation, 1520
Two-photon excitation of channelrhodopsin-2 at saturation, 1521
Two-Photon Imaging of Nonlinear Glutamate Release Dynamics at Bipolar Cell Synapses in the Mouse Retina, 1522
Two-photon optogenetics of dendritic spines and neural circuits, 1523
UBE3A, 1524
Ugurbil, 1525
Ulanovsky Lab, 1526
Ulrike Heberlein, 1527
Ultimate physical limits to computation, 1528
Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing, 1529
UNC Gene Therapy Center, 1530
Understanding transgenerational epigenetic inheritance via the gametes in mammals, 1531
Unique Identifiers for Authors, 1532
Unlimited Potential, Vanishing Opportunity, 1533
UPenn Vector Core, 1534
US 8257956 B2 - Sulfonylurea-responsive repressor proteins, 1535
USA, 1536
Using fast weights to deblur old memories, 1537
Using temperature to analyse temporal dynamics in the songbird motor pathway, 1538
UW Interactive Data Lab, 1539
Valentina Emiliani, 1540
Vantage Point by Richard Zare: My top 10 principles for success, 1541
Variation in cancer risk among tissues can be explained by the number of stem cell divisions, 1542
Vascular and Neurogenic Rejuvenation of the Aging Mouse Brain by Young Systemic Factors, 1543
ventral tegmental area, 1544
Vernor Vinge, 1545
Video game training enhances cognitive control in older adults, 1546
Villeda, 2011, 1547
Virally mediated optogenetic excitation and inhibition of pain in freely moving nontransgenic mice, 1548
Virginia Lee, 1549
Virginia Man-Yee Lee, 1550
visual cortex, 1551
Visualizing Hypothalamic Network Dynamics for Appetitive and Consummatory Behaviors, 1552
Visualizing mammalian brain area interactions by dual-axis two-photon calcium imaging, 1553
Visualizing Whole-Brain Activity and Development at the Single-Cell Level Using Light-Sheet Microscopy, 1554
Visually evoked activity in cortical cells imaged in freely moving animals, 1555
Vogelstein, 2014, 1556
voltage imaging, screening for better probes, 1557
Von Neumann architecture, 1558
Wandell, 1559
Wang, 2014, 1560
We Live in a Jungle of Artificial Intelligence that will Spawn Sentience, 1561
Web of Knowledge, 1562
well known lectures on physics, 1563
What Are The Most Cited Research Papers Of All Time?, 1564
What caused the flash crash? One big, bad trade, 1565
What errors do peer reviewers detect, and does training improve their ability to detect them?, 1566
What happens when a software bot goes on a darknet shopping spree?, 1567
What I Wish I Knew Before I Entered Grad School, 1568
What Is Life?, 1569
What Makes a Great Student in the Lab?, 1570
What the Frog’s Eye Tells the Frog’s Brain, 1571
When Are Results Too Good to Be True?, 1572
Who Reviews the Reviewers? Feasibility of Using a Fictitious Manuscript to Evaluate Peer Reviewer Performance, 1573
Who’s The Best In Drug Research? 22 Companies Ranked, 1574
whole animal 3D imaging, 1575
Whole-Brain Imaging with Single-Cell Resolution Using Chemical Cocktails and Computational Analysis, 1576
Why academics can^^e2^^80^^99t write, 1577
Why Academics Stink at Writing, 1578
Why Do Hubs Tend to Be Essential in Protein Networks?, 1579
Why Do We Feel Thirst? An Interview with Yuki Oka, 1580
Why I Am Not An Integrated Information Theorist (or, The Unconscious Expander), 1581
Why Incentive Plans Cannot Work, 1582
Why Is Academic Writing So Academic?, 1583
Why Most Published Research Findings Are False, 1584
Why nouns are learned before verbs: Linguistic relativity versus natural partitioning, 1585
Why optogenetics deserves the hype, 1586
Why physicists like models, and why biologists should, 1587
wikipathways, 1588
Wired article, 1589
Wireless magnetothermal deep brain stimulation, 1590
Wireless Neurosensor for Full-Spectrum Electrophysiology Recordings during Free Behavior, 1591
Wnt/Dkk Negative Feedback RegulatesSensory Organ Size in Zebra^^ef^^ac^^81sh, 1592
work at Intel and elsewhere on neuromorphic devices, 1593
Working Group on Data and Informatics, 1594
Working with Big Data in MATLAB, 1595, 1596
Wyss-Coray, 1597
Xcorr, 1598, 1599, 1600
xcorr, 1601
Xu, J Neuro 2008, 1602
Yael Maguire, 1603, 1604
Yin, 1605
Young blood reverses age-related impairments in cognitive function and synaptic plasticity in mice, 1606
Zador, 1607
Zhang, 1608
Zhang Lab, 1609
Zolpidem, 1610
Zolpidem Reduces Hippocampal Neuronal Activity in Freely Behaving Mice: A Large Scale Calcium Imaging Study with Miniaturized Fluorescence Microscope, 1611
zotero, 1612
Zuker lab publications, 1613